Trimeric structure of major outer membrane proteins homologous to OmpA in Porphyromonas gingivalis
- PMID: 15659668
- PMCID: PMC545718
- DOI: 10.1128/JB.187.3.902-911.2005
Trimeric structure of major outer membrane proteins homologous to OmpA in Porphyromonas gingivalis
Abstract
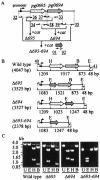
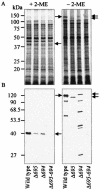
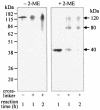
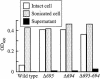
The major outer membrane proteins Pgm6 (41 kDa) and Pgm7 (40 kDa) of Porphyromonas gingivalis ATCC 33277 are encoded by open reading frames pg0695 and pg0694, respectively, which form a single operon. Pgm6 and Pgm7 (Pgm6/7) have a high degree of similarity to Escherichia coli OmpA in the C-terminal region and are predicted to form eight-stranded beta-barrels in the N-terminal region. By sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Pgm6/7 appear as bands with apparent molecular masses of 40 and 120 kDa, with and without a reducing agent, suggesting a monomer and trimer, respectively. To verify the predicted trimeric structure and function of Pgm6/7, we constructed three mutants with pg0695, pg0694, or both deleted. The double mutant produced no Pgm6/7. The single-deletion mutants appeared to contain less Pgm7 and Pgm6 and to form homotrimers that migrated slightly faster (115 kDa) and slower (130 kDa), respectively, than wild-type Pgm6/7 under nonreducing conditions. N-terminal amino acid sequencing and mass spectrometry analysis of partially digested Pgm6/7 detected only fragments from Pgm6 and Pgm7. Two-dimensional, diagonal electrophoresis and chemical cross-linking experiments with or without a reducing agent clearly showed that Pgm6/7 mainly form stable heterotrimers via intermolecular disulfide bonds. Furthermore, growth retardation and arrest of the three mutants and increased permeability of their outer membranes indicated that Pgm6/7 play an important role in outer membrane integrity. Based on results of liposome swelling experiments, these proteins are likely to function as a stabilizer of the cell wall rather than as a major porin in this organism.
Figures


Similar articles
-
Further evidence that major outer membrane proteins homologous to OmpA in Porphyromonas gingivalis stabilize bacterial cells.Oral Microbiol Immunol. 2007 Oct;22(5):356-60. doi: 10.1111/j.1399-302X.2007.00363.x. Oral Microbiol Immunol. 2007. PMID: 17803635
-
Identification and characterization of novel glycoproteins involved in growth and biofilm formation by Porphyromonas gingivalis.Mol Oral Microbiol. 2012 Dec;27(6):458-70. doi: 10.1111/j.2041-1014.2012.00659.x. Epub 2012 Jul 21. Mol Oral Microbiol. 2012. PMID: 23134611
-
Characterization of wheat germ agglutinin lectin-reactive glycosylated OmpA-like proteins derived from Porphyromonas gingivalis.Infect Immun. 2014 Nov;82(11):4563-71. doi: 10.1128/IAI.02069-14. Epub 2014 Aug 18. Infect Immun. 2014. PMID: 25135681 Free PMC article.
-
Identification of a gingipain-sensitive surface ligand of Porphyromonas gingivalis that induces Toll-like receptor 2- and 4-independent NF-kappaB activation in CHO cells.Infect Immun. 2009 Oct;77(10):4414-20. doi: 10.1128/IAI.00140-09. Epub 2009 Aug 10. Infect Immun. 2009. PMID: 19667049 Free PMC article.
-
Isolation and characterization of the Omp-PA porin from Porphyromonas asaccharolytica, determination of the omp-PA gene sequence and prediction of Omp-PA protein structure.Anaerobe. 2007 Apr;13(2):74-82. doi: 10.1016/j.anaerobe.2006.11.003. Epub 2007 Jan 16. Anaerobe. 2007. PMID: 17229581
Cited by
-
Evidence for a role of vertebrate Disp1 in long-range Shh signaling.Development. 2010 Jan;137(1):133-40. doi: 10.1242/dev.043547. Development. 2010. PMID: 20023168 Free PMC article.
-
Characterization of an OmpA-like outer membrane protein of the acidophilic iron-oxidizing bacterium, Acidithiobacillus ferrooxidans.Extremophiles. 2011 May;15(3):403-10. doi: 10.1007/s00792-011-0371-6. Epub 2011 Apr 7. Extremophiles. 2011. PMID: 21472537 Free PMC article.
-
Major membrane protein TDE2508 regulates adhesive potency in Treponema denticola.PLoS One. 2014 Feb 21;9(2):e89051. doi: 10.1371/journal.pone.0089051. eCollection 2014. PLoS One. 2014. PMID: 24586498 Free PMC article.
-
Selective sorting of cargo proteins into bacterial membrane vesicles.J Biol Chem. 2011 Jan 14;286(2):1269-76. doi: 10.1074/jbc.M110.185744. Epub 2010 Nov 5. J Biol Chem. 2011. PMID: 21056982 Free PMC article.
-
Characterization of Treponema denticola mutants defective in the major antigenic proteins, Msp and TmpC.PLoS One. 2014 Nov 17;9(11):e113565. doi: 10.1371/journal.pone.0113565. eCollection 2014. PLoS One. 2014. PMID: 25401769 Free PMC article.
References
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

