The Enterococcus faecalis sigV protein is an extracytoplasmic function sigma factor contributing to survival following heat, acid, and ethanol treatments
- PMID: 15659680
- PMCID: PMC545719
- DOI: 10.1128/JB.187.3.1022-1035.2005
The Enterococcus faecalis sigV protein is an extracytoplasmic function sigma factor contributing to survival following heat, acid, and ethanol treatments
Abstract
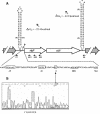
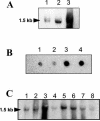
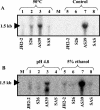
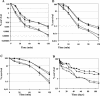
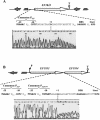
Analysis of the genome sequence of Enterococcus faecalis allowed the identification of two genes whose protein products showed 33 and 34% identity with those of sigV and yrhM of Bacillus subtilis, respectively. These genes, named sigV and rsiV, are predicted to encode members of the extracytoplasmic function subfamily of eubacterial RNA polymerase sigma and anti-sigma factors, respectively. This group of sigma factors has been shown to regulate gene expression in response to stress conditions. sigV and rsiV were shown to be under the control of the same promoter. The transcriptional start site was determined, and the 1.5-kb mRNA transcript was shown to be overexpressed under glucose and complete starvation, as well as under physicochemical treatments. Three mutants, affected in sigV, rsiV, and both genes, were constructed by double-crossover recombination within the genome of E. faecalis strain JH2-2. Compared with the wild type and the rsiV mutant, the sigV mutants were more susceptible to heat shock, acid, and ethanol treatments and displayed decreased survival during long-term starvation. A nisin-inducible sigV gene construction used in complementation assays restored the wild phenotype of the sigV mutants, confirming the involvement of SigV in the heat shock, ethanol, and acid stress responses. Northern blot analysis carried out with the three mutant strains revealed the inhibition of sigV expression by the related anti-sigma factor gene rsiV. In addition, putative candidates of the sigV regulon determined by computer search for the sigV promoter sequence were analyzed.
Figures




References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Batish, V. K., and B. Ranganathan. 1984. Occurrence of enterococci in milk and milk products. J. Dairy Sci. Technol. 19:189-195.
-
- Brutsche, S., and V. Braun. 1997. SigX of Bacillus subtilis replaces the ECF sigma factor FecI of Escherichia coli and is inhibited by RsiX. Mol. Gen. Genet. 256:416-425. - PubMed
-
- Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183-190. - PubMed
-
- Cao, M., P. A. Kobel, M. M. Morshedi, M. F. W. Wu, C. Paddon, and J. D. Helmann. 2002. Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, runoff transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316:443-457. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

