Potassium transport in a halophilic member of the bacteria domain: identification and characterization of the K+ uptake systems TrkH and TrkI from Halomonas elongata DSM 2581T
- PMID: 15659681
- PMCID: PMC545715
- DOI: 10.1128/JB.187.3.1036-1043.2005
Potassium transport in a halophilic member of the bacteria domain: identification and characterization of the K+ uptake systems TrkH and TrkI from Halomonas elongata DSM 2581T
Abstract
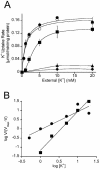
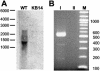
The halophilic bacterium Halomonas elongata accumulates K+, glutamate, and the compatible solute ectoine as osmoprotectants. By functional complementation of Escherichia coli mutants defective in K+ uptake, we cloned three genes that are required for K+ uptake in H. elongata. Two adjacent genes, named trkA (1,374 bp) and trkH (1,449 bp), were identified on an 8.5-kb DNA fragment, while a third gene, called trkI (1,479 bp), located at a different site in the H. elongata chromosome, was found on a second 8.5-kb fragment. The potential protein expressed by trkA is similar to the cytoplasmic NAD+/NADH binding protein TrkA from E. coli, which is required for the activity of the Trk K+ uptake system. The deduced amino acid sequences of trkH and trkI showed significant identity to the transmembrane protein of Trk transporters. K+ transport experiments with DeltatrkH and DeltatrkI mutants of H. elongata revealed that TrkI exhibits a Km value of 1.12 mM, while the TrkH system has a half-saturation constant of 3.36 mM. Strain KB12, relying on TrkH alone, accumulated K+ with a lower Vmax and required a higher K+ concentration for growth in highly saline medium than the wild type. Strain KB15, expressing only TrkI, showed the same phenotype and the same K+ transport kinetics as the wild type, proving that TrkI is the main K+ transport system in H. elongata. In the absence of both transporters TrkH and TrkI, K+ accumulation was not detectable. K+ transport was also abolished in a trkA deletion mutant, indicating that TrkI and TrkH depend on one type of TrkA protein. Reverse transcriptase PCR experiments and Northern hybridization analyses of the trkAH locus revealed cotranscription of trkAH as well as a monocistronic transcript with only trkA.
Figures

References
-
- Altendorf, K., M. Gaβel, W. Puppe, T. Möllenkamp, A. Zeeck, C. Boddien, K. Fendler, E. Bamberg, and S. Dröse. 1998. Structure and function of the Kdp-ATPase of Escherichia coli. Acta Physiol. Scand. 163(Suppl.):137-146. - PubMed
-
- Arahal, D. R., W. Ludwig, K. H. Schleifer, and A. Ventosa. 2002. Phylogeny of the family Halomonadaceae based on 23S and 165 rDNA sequence analyses. Int. J. Syst. Evol. Microbiol. 52:241-249. - PubMed
-
- Bossemeyer, D., A. Borchard, D. C. Dosch, G. C. Helmer, W. Epstein, I. R. Booth, and E. P. Bakker. 1989. K+-transport protein TrkA of Escherichia coli is a peripheral membrane protein that requires other trk gene products for attachment to the cytoplasmic membrane. J. Biol. Chem. 264:16403-16410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

