Clustered genes required for the synthesis of heterocyst envelope polysaccharide in Anabaena sp. strain PCC 7120
- PMID: 15659688
- PMCID: PMC545720
- DOI: 10.1128/JB.187.3.1114-1123.2005
Clustered genes required for the synthesis of heterocyst envelope polysaccharide in Anabaena sp. strain PCC 7120
Abstract
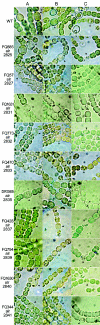
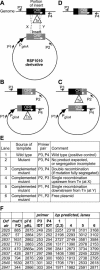
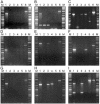
As demonstrated with alr2835 (hepA) and alr2834 (hepC) mutants, heterocysts of Anabaena sp. strain PCC 7120, a filamentous cyanobacterium, must have an envelope polysaccharide layer (the Hep+ phenotype) to fix dinitrogen in an oxygen-containing milieu (the Fox+ phenotype). Transpositions presumptively responsible for a Fox- phenotype were localized in open reading frames (ORFs) near hepA and hepC. A mutation in each of nine of these ORFs was complemented by a clone bearing only that single, intact ORF. Heterocysts of the nine mutants were found to lack an envelope polysaccharide layer. Complementation of mutations in alr2832 and alr2840 may have resulted from recombination. However, alr2825, alr2827, alr2831, alr2833, alr2837, alr2839, and alr2841, like hepA and hepC, are required for a Hep+ Fox+ phenotype.
Figures

Similar articles
-
Clustered genes required for synthesis and deposition of envelope glycolipids in Anabaena sp. strain PCC 7120.Mol Microbiol. 2005 Oct;58(1):227-43. doi: 10.1111/j.1365-2958.2005.04818.x. Mol Microbiol. 2005. PMID: 16164561
-
Alr5068, a Low-Molecular-Weight protein tyrosine phosphatase, is involved in formation of the heterocysts polysaccharide layer in the cyanobacterium Anabaena sp. PCC 7120.Res Microbiol. 2013 Oct;164(8):875-85. doi: 10.1016/j.resmic.2013.05.003. Epub 2013 Jul 1. Res Microbiol. 2013. PMID: 23827083
-
Genetic evidence that hepA gene is involved in the normal deposition of the envelope of both heterocysts and akinetes in Anabaena variabilis ATCC 29413.FEMS Microbiol Lett. 1994 Oct 15;123(1-2):63-7. doi: 10.1111/j.1574-6968.1994.tb07202.x. FEMS Microbiol Lett. 1994. PMID: 7988900
-
The cell wall in heterocyst formation by Anabaena sp. PCC 7120.J Basic Microbiol. 2009 Feb;49(1):5-24. doi: 10.1002/jobm.200800300. J Basic Microbiol. 2009. PMID: 19253332 Review.
-
Bacterial polysaccharide synthesis and gene nomenclature.Trends Microbiol. 1996 Dec;4(12):495-503. doi: 10.1016/s0966-842x(97)82912-5. Trends Microbiol. 1996. PMID: 9004408 Review.
Cited by
-
A TolC-like protein is required for heterocyst development in Anabaena sp. strain PCC 7120.J Bacteriol. 2007 Nov;189(21):7887-95. doi: 10.1128/JB.00750-07. Epub 2007 Aug 24. J Bacteriol. 2007. PMID: 17720784 Free PMC article.
-
Cell-specific gene expression in Anabaena variabilis grown phototrophically, mixotrophically, and heterotrophically.BMC Genomics. 2013 Nov 5;14(1):759. doi: 10.1186/1471-2164-14-759. BMC Genomics. 2013. PMID: 24191963 Free PMC article.
-
Impaired cell-cell communication in the multicellular cyanobacterium Anabaena affects carbon uptake, photosynthesis, and the cell wall.iScience. 2021 Jan 5;24(1):101977. doi: 10.1016/j.isci.2020.101977. eCollection 2021 Jan 22. iScience. 2021. PMID: 33458622 Free PMC article.
-
The composition of the global and feature specific cyanobacterial core-genomes.Front Microbiol. 2015 Mar 19;6:219. doi: 10.3389/fmicb.2015.00219. eCollection 2015. Front Microbiol. 2015. PMID: 25852675 Free PMC article.
-
Regulation by hetC of genes required for heterocyst differentiation and cell division in Anabaena sp. strain PCC 7120.J Bacteriol. 2005 Dec;187(24):8489-93. doi: 10.1128/JB.187.24.8489-8493.2005. J Bacteriol. 2005. PMID: 16321953 Free PMC article.
References
-
- Almon, H., and H. Böhme. 1980. Components and activity of the photosynthetic electron transport system of intact heterocysts isolated from the blue-green alga Nostoc muscorum. Biochim. Biophys. Acta 592:113-120. - PubMed
-
- Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

