Functional link between ribosome formation and biogenesis of iron-sulfur proteins
- PMID: 15660135
- PMCID: PMC548649
- DOI: 10.1038/sj.emboj.7600540
Functional link between ribosome formation and biogenesis of iron-sulfur proteins
Abstract
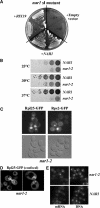
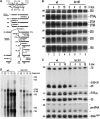
In genetic screens for ribosomal export mutants, we identified CFD1, NBP35 and NAR1 as factors involved in ribosome biogenesis. Notably, these components were recently reported to function in extramitochondrial iron-sulfur (Fe-S) cluster biosynthesis. In particular, Nar1 was implicated to generate the Fe-S clusters within Rli1, a potential substrate protein of unknown function. We tested whether the Fe-S protein Rli1 functions in ribosome formation. We report that rli1 mutants are impaired in pre-rRNA processing and defective in the export of both ribosomal subunits. In addition, Rli1p is associated with both pre-40S particles and mature 40S subunits, and with the eIF3 translation initiation factor complex. Our data reveal an unexpected link between ribosome biogenesis and the biosynthetic pathway of cytoplasmic Fe-S proteins.
Figures



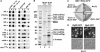
Similar articles
-
Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria.EMBO J. 2005 Feb 9;24(3):589-98. doi: 10.1038/sj.emboj.7600541. Epub 2005 Jan 20. EMBO J. 2005. PMID: 15660134 Free PMC article.
-
Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae.EMBO J. 2010 Jan 6;29(1):80-92. doi: 10.1038/emboj.2009.307. Epub 2009 Nov 5. EMBO J. 2010. PMID: 19893492 Free PMC article.
-
A bridging [4Fe-4S] cluster and nucleotide binding are essential for function of the Cfd1-Nbp35 complex as a scaffold in iron-sulfur protein maturation.J Biol Chem. 2012 Apr 6;287(15):12365-78. doi: 10.1074/jbc.M111.328914. Epub 2012 Feb 23. J Biol Chem. 2012. PMID: 22362766 Free PMC article.
-
Nuclear export and cytoplasmic maturation of ribosomal subunits.FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
-
Ribosome biogenesis in the yeast Saccharomyces cerevisiae.Genetics. 2013 Nov;195(3):643-81. doi: 10.1534/genetics.113.153197. Genetics. 2013. PMID: 24190922 Free PMC article. Review.
Cited by
-
Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis.Mol Cell Biol. 2008 Sep;28(18):5569-82. doi: 10.1128/MCB.00642-08. Epub 2008 Jul 14. Mol Cell Biol. 2008. PMID: 18625724 Free PMC article.
-
Fe-S cluster assembly in the supergroup Excavata.J Biol Inorg Chem. 2018 Jun;23(4):521-541. doi: 10.1007/s00775-018-1556-6. Epub 2018 Apr 5. J Biol Inorg Chem. 2018. PMID: 29623424 Free PMC article. Review.
-
ABCE1: A special factor that orchestrates translation at the crossroad between recycling and initiation.RNA Biol. 2017 Oct 3;14(10):1279-1285. doi: 10.1080/15476286.2016.1269993. Epub 2017 May 12. RNA Biol. 2017. PMID: 28498001 Free PMC article. Review.
-
Active regulator of SIRT1 is required for ribosome biogenesis and function.Nucleic Acids Res. 2013 Apr;41(7):4185-97. doi: 10.1093/nar/gkt129. Epub 2013 Mar 5. Nucleic Acids Res. 2013. PMID: 23462953 Free PMC article.
-
Structural basis of highly conserved ribosome recycling in eukaryotes and archaea.Nature. 2012 Feb 22;482(7386):501-6. doi: 10.1038/nature10829. Nature. 2012. PMID: 22358840 Free PMC article.
References
-
- Baßler J, Grandi P, Gadal O, Leßmann T, Tollervey D, Lechner J, Hurt EC (2001) Identification of a 60S pre-ribosomal particle that is closely linked to nuclear export. Mol Cell 8: 517–529 - PubMed
-
- Barton RM, Worman HJ (1999) Prenylated prelamin A interacts with Narf, a novel nuclear protein. J Biol Chem 274: 30008–30018 - PubMed
-
- Beinert H, Kiley PJ (1999) Fe–S proteins in sensing and regulatory functions. Curr Opin Chem Biol 3: 152–157 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

