An evaluation of detection methods for large lariat RNAs
- PMID: 15661842
- PMCID: PMC1370721
- DOI: 10.1261/rna.7124405
An evaluation of detection methods for large lariat RNAs
Abstract
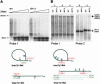
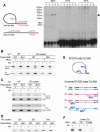
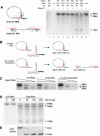
Ty1 elements are long terminal repeat (LTR) retrotransposons that reside within the genome of Saccharomyces cerevisiae. It has been known for many years that the 2'-5' phosphodiesterase Dbr1p, which debranches intron lariats, is required for efficient Ty1 transposition. A recent report suggested the intriguing possibility that Ty1 RNA forms a lariat as a transposition intermediate. We set out to further investigate the nature of the proposed Ty1 lariat branchpoint. However, using a wide range of techniques we were unable to find any evidence for the proposed lariat structure. Furthermore, we demonstrate that some of the techniques used in the initial study describing the lariat are capable of incorrectly reporting a lariat structure. Thus, the role of the Dbr1 protein in Ty1 retrotransposition remains elusive.
Figures

Similar articles
-
Relationship between RNA lariat debranching and Ty1 element retrotransposition.J Virol. 2003 Dec;77(23):12795-806. doi: 10.1128/jvi.77.23.12795-12806.2003. J Virol. 2003. PMID: 14610201 Free PMC article.
-
RNA branching and debranching in the yeast retrovirus-like element Ty1.Science. 2004 Jan 9;303(5655):240-3. doi: 10.1126/science.1087023. Science. 2004. PMID: 14716018
-
Saccharomyces cerevisiae RNA lariat debranching enzyme, Dbr1p, is required for completion of reverse transcription by the retrovirus-like element Ty1 and cleaves branched Ty1 RNAs.Mol Genet Genomics. 2021 Mar;296(2):409-422. doi: 10.1007/s00438-020-01753-y. Epub 2021 Jan 19. Mol Genet Genomics. 2021. PMID: 33464395
-
RNA Lariat Debranching Enzyme as a Retroviral and Long-Terminal-Repeat Retrotransposon Host Factor.Annu Rev Virol. 2020 Sep 29;7(1):189-202. doi: 10.1146/annurev-virology-012720-094902. Annu Rev Virol. 2020. PMID: 32991267 Review.
-
The Ty1 LTR-Retrotransposon of Budding Yeast, Saccharomyces cerevisiae.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0053-2014. doi: 10.1128/microbiolspec.MDNA3-0053-2014. Microbiol Spectr. 2015. PMID: 26104690 Review.
Cited by
-
Dual coding potential of a 2',5'-branched ribonucleotide in DNA.RNA. 2019 Jan;25(1):105-120. doi: 10.1261/rna.068486.118. Epub 2018 Oct 25. RNA. 2019. PMID: 30361268 Free PMC article.
-
hDbr1 is a nucleocytoplasmic shuttling protein with a protein phosphatase-like motif essential for debranching activity.Sci Rep. 2013;3:1090. doi: 10.1038/srep01090. Epub 2013 Jan 21. Sci Rep. 2013. PMID: 23346348 Free PMC article.
-
Unorthodox mRNA start site to extend the highly structured leader of retrotransposon Tto1 mRNA increases transposition rate.RNA. 2005 Aug;11(8):1181-91. doi: 10.1261/rna.2640105. RNA. 2005. PMID: 16043504 Free PMC article.
-
Function of a retrotransposon nucleocapsid protein.RNA Biol. 2010 Nov-Dec;7(6):642-54. doi: 10.4161/rna.7.6.14117. Epub 2010 Nov 1. RNA Biol. 2010. PMID: 21189452 Free PMC article. Review.
-
Structure-function analysis of yeast RNA debranching enzyme (Dbr1), a manganese-dependent phosphodiesterase.Nucleic Acids Res. 2005 Nov 7;33(19):6349-60. doi: 10.1093/nar/gki934. Print 2005. Nucleic Acids Res. 2005. PMID: 16275784 Free PMC article.
References
-
- Boeke, J.D., Garfinkel, D.J., Styles, C.A., and Fink, G.R. 1985. Ty elements transpose through an RNA intermediate. Cell 40: 491–500. - PubMed
-
- Chapman, K.B. and Boeke, J.D. 1991. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell 65: 483–492. - PubMed
-
- Chen, C. and Sarnow, P. 1995. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268: 415–417. - PubMed
-
- ———. 1998. Internal ribosome entry sites tests with circular mRNAs. Methods Mol. Biol. 77: 355–363. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
