U2AF binding selects for the high conservation of the C. elegans 3' splice site
- PMID: 15661845
- PMCID: PMC1370714
- DOI: 10.1261/rna.7221605
U2AF binding selects for the high conservation of the C. elegans 3' splice site
Abstract
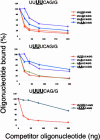
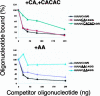
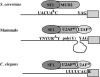
Caenorhabditis elegans is unusual among animals in having a highly conserved octamer sequence at the 3' splice site: UUUU CAG/R. This sequence can bind to the essential heterodimeric splicing factor U2AF, with U2AF65 contacting the U tract and U2AF35 contacting the splice site itself (AG/R). Here we demonstrate a strong correspondence between binding to U2AF of RNA oligonucleotides with variant octamer sequences and the frequency with which such variations occur in splice sites. C. elegans U2AF has a strong preference for the octamer sequence and exerts much of the pressure for 3' splice sites to have the precise UUUUCAG/R sequence. At two positions the splice site has a very strong preference for U even though alternative bases can also bind tightly to U2AF, suggesting that evolution can select against sequences that may have a relatively modest reduction in binding. Although pyrimidines are frequently present at the first base in the exon, U2AF has a very strong bias against them, arguing there is a mechanism to compensate for weakened U2AF binding at this position. Finally, the C in the consensus sequence must remain adjacent to the AG/R rather than to the stretch of U's, suggesting this C is recognized by U2AF35.
Figures


Similar articles
-
Both subunits of U2AF recognize the 3' splice site in Caenorhabditis elegans.Nature. 1999 Dec 16;402(6763):835-8. doi: 10.1038/45597. Nature. 1999. PMID: 10617207
-
Functional recognition of the 3' splice site AG by the splicing factor U2AF35.Nature. 1999 Dec 16;402(6763):832-5. doi: 10.1038/45590. Nature. 1999. PMID: 10617206
-
Cloning of Caenorhabditis U2AF65: an alternatively spliced RNA containing a novel exon.Mol Cell Biol. 1997 Feb;17(2):946-53. doi: 10.1128/MCB.17.2.946. Mol Cell Biol. 1997. PMID: 9001248 Free PMC article.
-
U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans.RNA. 1999 Apr;5(4):487-94. doi: 10.1017/s1355838299982225. RNA. 1999. PMID: 10199565 Free PMC article.
-
Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity.RNA. 1995 Mar;1(1):21-35. RNA. 1995. PMID: 7489484 Free PMC article.
Cited by
-
Caenorhabditis elegans operons contain a higher proportion of genes with multiple transcripts and use 3' splice sites differentially.PLoS One. 2010 Aug 27;5(8):e12456. doi: 10.1371/journal.pone.0012456. PLoS One. 2010. PMID: 20805997 Free PMC article.
-
mRNA Editing, Processing and Quality Control in Caenorhabditis elegans.Genetics. 2020 Jul;215(3):531-568. doi: 10.1534/genetics.119.301807. Genetics. 2020. PMID: 32632025 Free PMC article. Review.
-
The surveillance of pre-mRNA splicing is an early step in C. elegans RNAi of endogenous genes.Genes Dev. 2018 May 1;32(9-10):670-681. doi: 10.1101/gad.311514.118. Epub 2018 May 8. Genes Dev. 2018. PMID: 29739806 Free PMC article.
-
Branch point identification and sequence requirements for intron splicing in Plasmodium falciparum.Eukaryot Cell. 2011 Nov;10(11):1422-8. doi: 10.1128/EC.05193-11. Epub 2011 Sep 16. Eukaryot Cell. 2011. PMID: 21926333 Free PMC article.
-
U6 snRNA m6A modification is required for accurate and efficient splicing of C. elegans and human pre-mRNAs.Nucleic Acids Res. 2024 Aug 27;52(15):9139-9160. doi: 10.1093/nar/gkae447. Nucleic Acids Res. 2024. PMID: 38808663 Free PMC article.
References
-
- Berglund, J.A., Chua, K., Abovich, N., Reed, R., and Rosbash, M. 1997. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell 89: 781–787. - PubMed
-
- Blumenthal, T. and Steward, K. 1997. RNA processing and gene structure. In C. elegans II (ed. D.L. Riddle), pp. 117–145. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. - PubMed
-
- Brow, D.A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36: 333–360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
