Conserved spacing between the box C/D and C'/D' RNPs of the archaeal box C/D sRNP complex is required for efficient 2'-O-methylation of target RNAs
- PMID: 15661846
- PMCID: PMC1370718
- DOI: 10.1261/rna.7223405
Conserved spacing between the box C/D and C'/D' RNPs of the archaeal box C/D sRNP complex is required for efficient 2'-O-methylation of target RNAs
Abstract
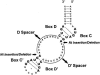
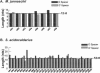
RNA-guided nucleotide modification complexes direct the post-transcriptional nucleotide modification of both archaeal and eukaryotic RNAs. We have previously demonstrated that efficient 2'-O-methylation activity guided by an in vitro reconstituted archaeal box C/D sRNP requires juxtaposed box C/D and C'/D' RNP complexes. In these experiments, we investigate the importance of spatially positioning the box C/D and C'/D' RNPs within the sRNP complex for nucleotide modification. Initial sequence analysis of 245 archaeal box C/D sRNAs from both Eukyarchaeota and Crenarchaeota kingdoms revealed highly conserved spacing between the box C/D and C'/D' RNA motifs. Distances between boxes C to D' and C' to D (D' and D spacers, respectively) exhibit highly constrained lengths of 12 nucleotides (nt). Methanocaldococcus jannaschii sR8 sRNA, a model box C/D sRNA with D and D' spacers of 12 nt, was mutated to alter the distance between the two RNA motifs. sRNAs with longer or shorter spacer regions could still form sRNPs by associating with box C/D core proteins, L7, Nop56/58, and fibrillarin, comparable to wild-type sR8. However, these reconstituted box C/D sRNP complexes were severely deficient in methylation activity. Alteration of the D and D' spacer lengths disrupted the guided methylation activity of both the box C/D and C'/D' RNP complexes. When only one spacer region was altered, methylation activity of the corresponding RNP was lost. Collectively, these results demonstrate the importance of box C/D and C'/D' RNP positioning for preservation of critical inter-RNP interactions required for efficient box C/D sRNP-guided nucleotide methylation.
Figures
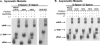
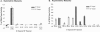
Similar articles
-
Efficient RNA 2'-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C'/D' RNPs.EMBO J. 2003 Aug 1;22(15):3930-40. doi: 10.1093/emboj/cdg368. EMBO J. 2003. PMID: 12881427 Free PMC article.
-
The coiled-coil domain of the Nop56/58 core protein is dispensable for sRNP assembly but is critical for archaeal box C/D sRNP-guided nucleotide methylation.RNA. 2006 Jun;12(6):1092-103. doi: 10.1261/rna.2230106. Epub 2006 Apr 6. RNA. 2006. PMID: 16601205 Free PMC article.
-
Structural features of the guide:target RNA duplex required for archaeal box C/D sRNA-guided nucleotide 2'-O-methylation.RNA. 2007 Jun;13(6):899-911. doi: 10.1261/rna.517307. Epub 2007 Apr 16. RNA. 2007. PMID: 17438123 Free PMC article.
-
In vitro reconstitution and affinity purification of catalytically active archaeal box C/D sRNP complexes.Methods Enzymol. 2007;425:263-82. doi: 10.1016/S0076-6879(07)25012-8. Methods Enzymol. 2007. PMID: 17673088 Review.
-
RNA structure and function in C/D and H/ACA s(no)RNPs.Curr Opin Struct Biol. 2004 Jun;14(3):335-43. doi: 10.1016/j.sbi.2004.05.006. Curr Opin Struct Biol. 2004. PMID: 15193314 Review.
Cited by
-
Emerging Data on the Diversity of Molecular Mechanisms Involving C/D snoRNAs.Noncoding RNA. 2021 May 6;7(2):30. doi: 10.3390/ncrna7020030. Noncoding RNA. 2021. PMID: 34066559 Free PMC article. Review.
-
The roles of NOP56 in cancer and SCA36.Pathol Oncol Res. 2023 Jan 19;29:1610884. doi: 10.3389/pore.2023.1610884. eCollection 2023. Pathol Oncol Res. 2023. PMID: 36741964 Free PMC article. Review.
-
High-resolution structure of eukaryotic Fibrillarin interacting with Nop56 amino-terminal domain.RNA. 2021 Apr;27(4):496-512. doi: 10.1261/rna.077396.120. Epub 2021 Jan 22. RNA. 2021. PMID: 33483369 Free PMC article.
-
Analysis of a critical interaction within the archaeal box C/D small ribonucleoprotein complex.J Biol Chem. 2009 May 29;284(22):15317-24. doi: 10.1074/jbc.M901368200. Epub 2009 Mar 31. J Biol Chem. 2009. PMID: 19336398 Free PMC article.
-
The spatial-functional coupling of box C/D and C'/D' RNPs is an evolutionarily conserved feature of the eukaryotic box C/D snoRNP nucleotide modification complex.Mol Cell Biol. 2011 Jan;31(2):365-74. doi: 10.1128/MCB.00918-10. Epub 2010 Nov 1. Mol Cell Biol. 2011. PMID: 21041475 Free PMC article.
References
-
- Aittaleb, M., Rashid, R., Chen, Q., Palmer, J., Daniels, C., and Li, H. 2003. Structure and function of archaeal box C/D sRNP core proteins. Nat. Struct. Biol. 10: 256–263. - PubMed
-
- Bachellerie, J.P., Cavaille, J., and Huttenhofer, A. 2002. The expanding snoRNA world. Biochimie 84: 775–790. - PubMed
-
- Balakin, A., Smith, L., and Fournier, M.J. 1996. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 86: 823–834. - PubMed
-
- Borovjagin, A. and Gerbi, S. 1999. U3 small nucleolar RNA is essential for cleavage at sites 1, 2, and 3 in pre-rRNA and determines which rRNA processing pathway is taken in Xenopus oocytes. J. Mol. Biol. 286: 1347–1363. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
