H3N2 influenza virus transmission from swine to turkeys, United States
- PMID: 15663853
- PMCID: PMC3323362
- DOI: 10.3201/eid1012.040581
H3N2 influenza virus transmission from swine to turkeys, United States
Abstract
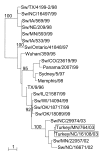
In 1998, a novel H3N2 reassortant virus emerged in the United States swine population. We report the interspecies transmission of this virus to turkeys in two geographically distant farms in the United States in 2003. This event is of concern, considering the reassortment capacity of this virus and the susceptibility of turkey to infection by avian influenza viruses. Two H3N2 isolates, A/turkey/NC/16108/03 and A/turkey/MN/764/03, had 98.0% to 99.9% nucleotide sequence identity to each other in all eight gene segments. All protein components of the turkey isolates had 97% to 98% sequence identity to swine H3N2 viruses, thus demonstrating interspecies transmission from pigs to turkeys. The turkey isolates were better adapted to avian hosts than were their closest swine counterparts, which suggests that the viruses had already begun to evolve in the new host. The isolation of swine-like H3N2 influenza viruses from turkeys raises new concerns for the generation of novel viruses that could affect humans.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
