Fast and reliable prediction of noncoding RNAs
- PMID: 15665081
- PMCID: PMC548974
- DOI: 10.1073/pnas.0409169102
Fast and reliable prediction of noncoding RNAs
Abstract
We report an efficient method for detecting functional RNAs. The approach, which combines comparative sequence analysis and structure prediction, already has yielded excellent results for a small number of aligned sequences and is suitable for large-scale genomic screens. It consists of two basic components: (i) a measure for RNA secondary structure conservation based on computing a consensus secondary structure, and (ii) a measure for thermodynamic stability, which, in the spirit of a z score, is normalized with respect to both sequence length and base composition but can be calculated without sampling from shuffled sequences. Functional RNA secondary structures can be identified in multiple sequence alignments with high sensitivity and high specificity. We demonstrate that this approach is not only much more accurate than previous methods but also significantly faster. The method is implemented in the program rnaz, which can be downloaded from www.tbi.univie.ac.at/~wash/RNAz. We screened all alignments of length n > or = 50 in the Comparative Regulatory Genomics database, which compiles conserved noncoding elements in upstream regions of orthologous genes from human, mouse, rat, Fugu, and zebrafish. We recovered all of the known noncoding RNAs and cis-acting elements with high significance and found compelling evidence for many other conserved RNA secondary structures not described so far to our knowledge.
Figures
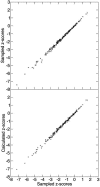
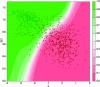
Comment in
-
Tracking down noncoding RNAs.Proc Natl Acad Sci U S A. 2005 Feb 15;102(7):2269-70. doi: 10.1073/pnas.0500129102. Epub 2005 Feb 9. Proc Natl Acad Sci U S A. 2005. PMID: 15703286 Free PMC article. No abstract available.
Similar articles
-
RNAz 2.0: improved noncoding RNA detection.Pac Symp Biocomput. 2010:69-79. Pac Symp Biocomput. 2010. PMID: 19908359
-
Identifying structural noncoding RNAs using RNAz.Curr Protoc Bioinformatics. 2007 Sep;Chapter 12:Unit 12.7. doi: 10.1002/0471250953.bi1207s19. Curr Protoc Bioinformatics. 2007. PMID: 18428784
-
The RNAz web server: prediction of thermodynamically stable and evolutionarily conserved RNA structures.Nucleic Acids Res. 2007 Jul;35(Web Server issue):W335-8. doi: 10.1093/nar/gkm222. Epub 2007 Apr 22. Nucleic Acids Res. 2007. PMID: 17452347 Free PMC article.
-
Sequence and structure analysis of noncoding RNAs.Methods Mol Biol. 2010;609:285-306. doi: 10.1007/978-1-60327-241-4_17. Methods Mol Biol. 2010. PMID: 20221926 Review.
-
Folding and finding RNA secondary structure.Cold Spring Harb Perspect Biol. 2010 Dec;2(12):a003665. doi: 10.1101/cshperspect.a003665. Epub 2010 Aug 4. Cold Spring Harb Perspect Biol. 2010. PMID: 20685845 Free PMC article. Review.
Cited by
-
Testing the nearest neighbor model for canonical RNA base pairs: revision of GU parameters.Biochemistry. 2012 Apr 24;51(16):3508-22. doi: 10.1021/bi3002709. Epub 2012 Apr 10. Biochemistry. 2012. PMID: 22490167 Free PMC article.
-
Transcripts with in silico predicted RNA structure are enriched everywhere in the mouse brain.BMC Genomics. 2012 May 31;13:214. doi: 10.1186/1471-2164-13-214. BMC Genomics. 2012. PMID: 22651826 Free PMC article.
-
Comparison of splice sites reveals that long noncoding RNAs are evolutionarily well conserved.RNA. 2015 May;21(5):801-12. doi: 10.1261/rna.046342.114. Epub 2015 Mar 23. RNA. 2015. PMID: 25802408 Free PMC article.
-
An ancient repeat sequence in the ATP synthase beta-subunit gene of forcipulate sea stars.J Mol Evol. 2007 Nov;65(5):564-73. doi: 10.1007/s00239-007-9036-6. Epub 2007 Oct 2. J Mol Evol. 2007. PMID: 17909692
-
Regulatory context drives conservation of glycine riboswitch aptamers.PLoS Comput Biol. 2019 Dec 20;15(12):e1007564. doi: 10.1371/journal.pcbi.1007564. eCollection 2019 Dec. PLoS Comput Biol. 2019. PMID: 31860665 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

