An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs
- PMID: 15665103
- PMCID: PMC547832
- DOI: 10.1073/pnas.0406347102
An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs
Abstract
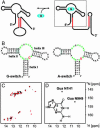
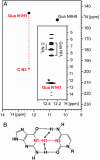
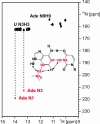
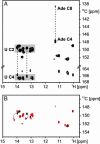
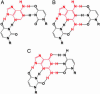
Riboswitches are highly structured RNA elements that control the expression of many bacterial genes by binding directly to small metabolite molecules with high specificity and affinity. In Bacillus subtilis, two classes of riboswitches have been described that discriminate between guanine and adenine despite an extremely high degree of homology both in their primary and secondary structure. We have identified intermolecular base triples between both purine ligands and their respective riboswitch RNAs by NMR spectroscopy. Here, specificity is mediated by the formation of a Watson-Crick base pair between the guanine ligand and a C residue or the adenine ligand and a U residue of the cognate riboswitch RNA, respectively. In addition, a second base-pairing interaction common to both riboswitch purine complexes involves a uridine residue of the RNA and the N3/N9 edge of the purine ligands. This base pairing is mediated by a previously undescribed hydrogen-bonding scheme that contributes to the affinity of the RNA-ligand interaction. The observed intermolecular hydrogen bonds between the purine ligands and the RNA rationalize the previously observed change in specificity upon a C to U mutation in the core of the purine riboswitch RNAs and the differences in the binding affinities for a number of purine analogs.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

