Functional genomics of the regulation of the nitrate assimilation pathway in Chlamydomonas
- PMID: 15665251
- PMCID: PMC1065353
- DOI: 10.1104/pp.104.050914
Functional genomics of the regulation of the nitrate assimilation pathway in Chlamydomonas
Abstract
The existence of mutants at specific steps in a pathway is a valuable tool of functional genomics in an organism. Heterologous integration occurring during transformation with a selectable marker in Chlamydomonas (Chlamydomonas reinhardtii) has been used to generate an ordered mutant library. A strain, having a chimeric construct (pNia1::arylsulfatase gene) as a sensor of the Nia1 gene promoter activity, was transformed with a plasmid bearing the paramomycin resistance AphVIII gene to generate insertional mutants defective at regulatory steps of the nitrate assimilation pathway. Twenty-two thousand transformants were obtained and maintained in pools of 96 for further use. The mutant library was screened for the following phenotypes: insensitivity to the negative signal of ammonium, insensitivity to the positive signal of nitrate, overexpression in nitrate, and inability to use nitrate. Analyses of mutants showed that (1) the number or integrated copies of the gene marker is close to 1; (2) the probability of cloning the DNA region at the marker insertion site is high (76%); (3) insertions occur randomly; and (4) integrations at different positions and orientations of the same genomic region appeared in at least three cases. Some of the mutants analyzed were found to be affected at putative new genes related to regulatory functions, such as guanylate cyclase, protein kinase, peptidyl-prolyl isomerase, or DNA binding. The Chlamydomonas mutant library constructed would also be valuable to identify any other gene with a screenable phenotype.
Figures
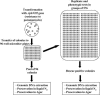

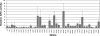


References
-
- Cenkci B, Petersen JL, Small GD (2003) REX1, a novel gene required for DNA repair. J Biol Chem 278: 22574–22577 - PubMed
-
- Colombo SL, Pollock SV, Eger KA, Godfrey AC, Adams JE, Mason CB, Moroney JV (2002) Use of the bleomycin resistance gene to generate tagged insertional mutants of Chlamydomonas reinhardtii that require elevated CO2 for optimal growth. Funct Plant Biol 29: 231–241 - PubMed
-
- Crawford NM, Arst HN Jr (1993) The molecular genetics of nitrate assimilation in fungi and plants. Annu Rev Genet 27: 115–146 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

