Recognizing a single base in an individual DNA strand: a step toward DNA sequencing in nanopores
- PMID: 15666419
- PMCID: PMC1828035
- DOI: 10.1002/anie.200462114
Recognizing a single base in an individual DNA strand: a step toward DNA sequencing in nanopores
Abstract
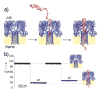
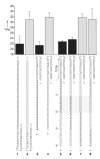
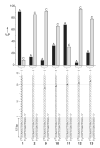
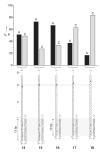
Functional supramolecular chemistry at the single-molecule level. Single strands of DNA can be captured inside α-hemolysin transmembrane pore protein to form single-species α-HL·DNA pseudorotaxanes. This process can be used to identify a single adenine nucleotide at a specific location on a strand of DNA by the characteristic reductions in the α-HL ion conductance. This study suggests that α-HL-mediated single-molecule DNA sequencing might be fundamentally feasible.
Figures
References
-
- Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Science. 1996;274:1859. - PubMed
-
- Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proc Natl Acad Sci USA. 1996;93:13770. - PMC - PubMed
- Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Biophys J. 1999;77:3227. - PMC - PubMed
- Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Proc Natl Acad Sci USA. 2000;97:1079. - PMC - PubMed
- Howorka S, Cheley S, Bayley H. Nat Biotechnol. 2001;19:636. - PubMed
-
- Jirage KB, Hulteen JC, Martin CR. Science. 1997;278:655.
- Schmidt C, Mayer M, Vogel H. Angew Chem. 2000;112:3267. Angew. Chem. Int. Ed. 2000, 39, 3137. - PubMed
- Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. Nature. 2001;412:166. - PubMed
- Bayley H, Cremer PS. Nature. 2001;413:226. - PubMed
- Chen P, Mitsui T, Farmer DB, Golovchenko J, Gordon RG, Branton D. Nano Lett. 2004;4:1333. - PMC - PubMed
- Chang H, Kosari F, Andreadakis G, Alam MA, Vasmatzis G, Bashir R. Nano Lett. 2004;4:1551.
-
- Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Phys Rev Lett. 2003;90:238101. - PubMed
- Sanchez-Quesada J, Saghatelian A, Cheley S, Bayley H, Ghadiri MR. Angew Chem Int Ed. 2004;43:3063. Angew. Chem. 2004, 116, 3125. - PMC - PubMed
- Nakane J, Wiggin M, Marziali A. Biophys J. 2004;87:615. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

