Nibrin functions in Ig class-switch recombination
- PMID: 15668383
- PMCID: PMC547877
- DOI: 10.1073/pnas.0409191102
Nibrin functions in Ig class-switch recombination
Abstract
Nijmegen breakage syndrome (NBS) is a rare autosomal recessive disorder characterized by predisposition to hematopoietic malignancy, cell-cycle checkpoint defects, and ionizing radiation sensitivity. NBS is caused by a hypomorphic mutation of the NBS1 gene, encoding nibrin, which forms a protein complex with Mre11 and Rad50, both involved in DNA repair. Nibrin localizes to chromosomal sites of class switching, and B cells from NBS patients show an enhanced presence of microhomologies at the sites of switch recombination. Because nibrin is crucial for embryonic survival, direct demonstration by targeted deletion that nibrin functions in class switch recombination has been lacking. Here, we show by cell-type-specific conditional inactivation of Nbn, the murine homologue of NBS1, that nibrin plays a role in the repair of gamma-irradiation damage, maintenance of chromosomal stability, and the recombination of Ig constant region genes in B lymphocytes.
Figures
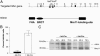
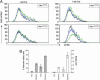
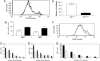
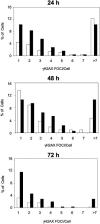
References
-
- Varon, R., Vissinga, C., Platzer, M., Cerosaletti, K. M., Chrzanowska, K. H., Saar, K., Beckmann, G., Seemanova, E., Cooper, P. R., Nowak, N. J., et al. (1998) Cell 93, 467-476. - PubMed
-
- Tauchi, H., Kobayashi, J., Morishima, K., van Gent, D. C., Shiraishi, T., Verkaik, N. S., vanHeems, D., Ito, E., Nakamura, A., Sonoda, E., et al. (2002) Nature 420, 93-98. - PubMed
-
- Zhu, J., Petersen, S., Tessarollo, L. & Nussenzweig, A. (2001) Curr. Biol. 11, 105-109. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

