Partitioning between unfolding and release of native domains during ClpXP degradation determines substrate selectivity and partial processing
- PMID: 15671177
- PMCID: PMC547888
- DOI: 10.1073/pnas.0409634102
Partitioning between unfolding and release of native domains during ClpXP degradation determines substrate selectivity and partial processing
Abstract
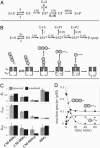
Energy-dependent proteases, such as ClpXP, are responsible for the regulated destruction of proteins in all cells. AAA+ ATPases in these proteases bind protein substrates and power their mechanical denaturation and subsequent translocation into a secluded degradation chamber where polypeptide cleavage occurs. Here, we show that model unfolded substrates are engaged rapidly by ClpXP and are then spooled into the degradation chamber at a rate proportional to their length. Degradation and competition studies indicate that ClpXP initially binds native and unfolded substrates similarly. However, stable native substrates then partition between frequent release and infrequent denaturation, with only the latter step resulting in committed degradation. During degradation of a fusion protein with three tandem native domains, partially degraded species with one and two intact domains accumulated. These processed proteins were not bound to the enzyme, showing that release can occur even after translocation and degradation of a substrate have commenced. The release of stable substrates and committed engagement of denatured or unstable native molecules ensures that ClpXP degrades less stable substrates in a population preferentially. This mechanism prevents trapping of the enzyme in futile degradation attempts and ensures that the energy of ATP hydrolysis is used efficiently for protein degradation.
Figures


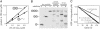
Similar articles
-
Single-molecule denaturation and degradation of proteins by the AAA+ ClpXP protease.Proc Natl Acad Sci U S A. 2009 Nov 17;106(46):19340-5. doi: 10.1073/pnas.0910484106. Epub 2009 Nov 5. Proc Natl Acad Sci U S A. 2009. PMID: 19892734 Free PMC article.
-
Multistep substrate binding and engagement by the AAA+ ClpXP protease.Proc Natl Acad Sci U S A. 2020 Nov 10;117(45):28005-28013. doi: 10.1073/pnas.2010804117. Epub 2020 Oct 26. Proc Natl Acad Sci U S A. 2020. PMID: 33106413 Free PMC article.
-
Protein unfolding by a AAA+ protease is dependent on ATP-hydrolysis rates and substrate energy landscapes.Nat Struct Mol Biol. 2008 Feb;15(2):139-45. doi: 10.1038/nsmb.1380. Epub 2008 Jan 27. Nat Struct Mol Biol. 2008. PMID: 18223658
-
ClpXP, an ATP-powered unfolding and protein-degradation machine.Biochim Biophys Acta. 2012 Jan;1823(1):15-28. doi: 10.1016/j.bbamcr.2011.06.007. Epub 2011 Jun 27. Biochim Biophys Acta. 2012. PMID: 21736903 Free PMC article. Review.
-
ClpP: a structurally dynamic protease regulated by AAA+ proteins.J Struct Biol. 2012 Aug;179(2):202-10. doi: 10.1016/j.jsb.2012.05.003. Epub 2012 May 14. J Struct Biol. 2012. PMID: 22595189 Review.
Cited by
-
Antibacterial activity of and resistance to small molecule inhibitors of the ClpP peptidase.ACS Chem Biol. 2013 Dec 20;8(12):2669-77. doi: 10.1021/cb400577b. Epub 2013 Oct 4. ACS Chem Biol. 2013. PMID: 24047344 Free PMC article.
-
Dependence of proteasome processing rate on substrate unfolding.J Biol Chem. 2011 May 20;286(20):17495-502. doi: 10.1074/jbc.M110.212027. Epub 2011 Mar 28. J Biol Chem. 2011. PMID: 21454622 Free PMC article.
-
Dissection of Axial-Pore Loop Function during Unfolding and Translocation by a AAA+ Proteolytic Machine.Cell Rep. 2015 Aug 11;12(6):1032-41. doi: 10.1016/j.celrep.2015.07.007. Epub 2015 Jul 30. Cell Rep. 2015. PMID: 26235618 Free PMC article.
-
Protein unfolding and degradation by the AAA+ Lon protease.Protein Sci. 2012 Feb;21(2):268-78. doi: 10.1002/pro.2013. Epub 2012 Jan 4. Protein Sci. 2012. PMID: 22162032 Free PMC article.
-
Roles of the ClpX IGF loops in ClpP association, dissociation, and protein degradation.Protein Sci. 2019 Apr;28(4):756-765. doi: 10.1002/pro.3590. Epub 2019 Mar 4. Protein Sci. 2019. PMID: 30767302 Free PMC article.
References
-
- Neuwald, A. F., Aravind, L., Spouge, J. L. & Koonin, E. V. (1999) Genome Res. 9, 27-43. - PubMed
-
- Kim, Y. I., Burton, R. E., Burton, B. M., Sauer, R. T. & Baker, T. A. (2000) Mol. Cell 5, 639-648. - PubMed
-
- Kenniston, J. A., Baker, T. A., Fernandez, J. M. & Sauer, R. T. (2003) Cell 114, 511-520. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

