In vitro synthesis of uniform poly(dG)-poly(dC) by Klenow exo- fragment of polymerase I
- PMID: 15673713
- PMCID: PMC548336
- DOI: 10.1093/nar/gki178
In vitro synthesis of uniform poly(dG)-poly(dC) by Klenow exo- fragment of polymerase I
Abstract
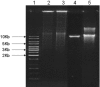
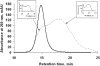
In this paper, we describe a production procedure of the one-to-one double helical complex of poly(dG)-poly(dC), characterized by a well-defined length (up to 10 kb) and narrow size distribution of molecules. Direct evidence of strands slippage during poly(dG)-poly(dC) synthesis by Klenow exo(-) fragment of polymerase I is obtained by fluorescence resonance energy transfer (FRET). We show that the polymer extension results in an increase in the separation distance between fluorescent dyes attached to 5' ends of the strands in time and, as a result, losing communication between the dyes via FRET. Analysis of the products of the early steps of the synthesis by high-performance liquid chromatography and mass spectroscopy suggest that only one nucleotide is added to each of the strand composing poly(dG)-poly(dC) in the elementary step of the polymer extension. We show that proper pairing of a base at the 3' end of the primer strand with a base in sequence of the template strand is required for initiation of the synthesis. If the 3' end nucleotide in either poly(dG) or poly(dC) strand is substituted for A, the polymer does not grow. Introduction of the T-nucleotide into the complementary strand to permit pairing with A-nucleotide results in the restoration of the synthesis. The data reported here correspond with a slippage model of replication, which includes the formation of loops on the 3' ends of both strands composing poly(dG)-poly(dC) and their migration over long-molecular distances (microm) to 5' ends of the strands.
Figures


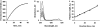




Similar articles
-
Synthesis of novel poly(dG)-poly(dG)-poly(dC) triplex structure by Klenow exo- fragment of DNA polymerase I.Nucleic Acids Res. 2005 Nov 27;33(20):6515-21. doi: 10.1093/nar/gki963. Print 2005. Nucleic Acids Res. 2005. PMID: 16314313 Free PMC article.
-
Synthesis of DNA-Based Nanowires.Methods Mol Biol. 2018;1811:23-47. doi: 10.1007/978-1-4939-8582-1_3. Methods Mol Biol. 2018. PMID: 29926444
-
In vitro bypass of malondialdehyde-deoxyguanosine adducts: differential base selection during extension by the Klenow fragment of DNA polymerase I is the critical determinant of replication outcome.Biochemistry. 2004 Sep 21;43(37):11828-35. doi: 10.1021/bi049360f. Biochemistry. 2004. PMID: 15362868
-
Synthesis of long DNA-based nanowires.Methods Mol Biol. 2011;749:115-40. doi: 10.1007/978-1-61779-142-0_9. Methods Mol Biol. 2011. PMID: 21674369
-
Conformational variability of poly(dA-dT).poly(dA-dT) and some other deoxyribonucleic acids includes a novel type of double helix.J Biomol Struct Dyn. 1985 Aug;3(1):67-83. doi: 10.1080/07391102.1985.10508399. J Biomol Struct Dyn. 1985. PMID: 3917211 Review.
Cited by
-
Ag+-Mediated Folding of Long Polyguanine Strands to Double and Quadruple Helixes.Nanomaterials (Basel). 2024 Apr 11;14(8):663. doi: 10.3390/nano14080663. Nanomaterials (Basel). 2024. PMID: 38668157 Free PMC article.
-
Mono and trimethine cyanines Cyan 40 and Cyan 2 as probes for highly selective fluorescent detection of non-canonical DNA structures.J Fluoresc. 2011 Jan;21(1):223-30. doi: 10.1007/s10895-010-0709-y. Epub 2010 Sep 1. J Fluoresc. 2011. PMID: 20809136
-
Assembling of G-strands into novel tetra-molecular parallel G4-DNA nanostructures using avidin-biotin recognition.Nucleic Acids Res. 2008 Sep;36(15):5050-60. doi: 10.1093/nar/gkn459. Epub 2008 Jul 28. Nucleic Acids Res. 2008. PMID: 18663013 Free PMC article.
-
Fluorescent Silver Nanoclusters Associated with Double-Stranded Poly(dGdC) DNA.Nanomaterials (Basel). 2025 Mar 5;15(5):397. doi: 10.3390/nano15050397. Nanomaterials (Basel). 2025. PMID: 40072200 Free PMC article.
-
Synthesis of novel poly(dG)-poly(dG)-poly(dC) triplex structure by Klenow exo- fragment of DNA polymerase I.Nucleic Acids Res. 2005 Nov 27;33(20):6515-21. doi: 10.1093/nar/gki963. Print 2005. Nucleic Acids Res. 2005. PMID: 16314313 Free PMC article.
References
-
- Kornberg A. DNA Synthesis. San Francisco, CA: W. J. Freeman and Co.; 1974. DNA polymerase of E.coli. p. 103.
-
- Schachman H.K., Adler J., Radding C.M., Lehman I.R., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. VII. Synthesis of a polymer of deoxyadenylate and deoxythymidylate. J. Biol. Chem. 1960;235:3242–3249. - PubMed
-
- Radding C.M., Josse J., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XII. A polymer of deoxyguanylate and deoxycytidylate. J. Biol. Chem. 1962;237:2869–2876. - PubMed
-
- Inman R.B., Baldwin R.L. Helix-random coil transitions in DNA homopolymer pairs. J. Mol. Biol. 1964;41:452–469. - PubMed
-
- Burd J.F., Wells R.D. Effect of incubation conditions on the nucleotide sequence of DNA products of unprimed DNA polymerase reactions. J. Mol. Biol. 1970;53:435–459. - PubMed

