Conservation of amino acids in human rhinovirus 3C protease correlates with broad-spectrum antiviral activity of rupintrivir, a novel human rhinovirus 3C protease inhibitor
- PMID: 15673742
- PMCID: PMC547258
- DOI: 10.1128/AAC.49.2.619-626.2005
Conservation of amino acids in human rhinovirus 3C protease correlates with broad-spectrum antiviral activity of rupintrivir, a novel human rhinovirus 3C protease inhibitor
Abstract
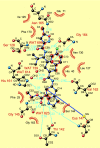
The picornavirus 3C protease is required for the majority of proteolytic cleavages that occur during the viral life cycle. Comparisons of published amino acid sequences from 6 human rhinoviruses (HRV) and 20 human enteroviruses (HEV) show considerable variability in the 3C protease-coding region but strict conservation of the catalytic triad residues. Rupintrivir (formerly AG7088) is an irreversible inhibitor of HRV 3C protease with potent in vitro activity against all HRV serotypes (48 of 48), HEV strains (4 of 4), and untyped HRV field isolates (46 of 46) tested. To better understand the relationship between in vitro antiviral activity and 3C protease-rupintrivir binding interactions, we performed nucleotide sequence analyses on an additional 21 HRV serotypes and 11 HRV clinical isolates. Antiviral activity was also determined for 23 HRV clinical isolates and four additional HEV strains. Sequence comparison of 3C proteases (n = 58) show that 13 and 11 of the 14 amino acids that are involved in side chain interactions with rupintrivir are strictly conserved among HRV and HEV, respectively. These sequence analyses are consistent with the comparable in vitro antiviral potencies of rupintrivir against all HRV serotypes, HRV isolates, and HEV strains tested (50% effective concentration range, 3 to 183 nM; n = 125). In summary, the conservation of critical amino acid residues in 3C protease and the observation of potent, broad-spectrum antipicornavirus activity of rupintrivir highlight the advantages of 3C protease as an antiviral target.
Figures



Similar articles
-
In vitro resistance study of rupintrivir, a novel inhibitor of human rhinovirus 3C protease.Antimicrob Agents Chemother. 2007 Dec;51(12):4366-73. doi: 10.1128/AAC.00905-07. Epub 2007 Oct 1. Antimicrob Agents Chemother. 2007. PMID: 17908951 Free PMC article.
-
In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease.Antimicrob Agents Chemother. 1999 Oct;43(10):2444-50. doi: 10.1128/AAC.43.10.2444. Antimicrob Agents Chemother. 1999. PMID: 10508022 Free PMC article.
-
In vitro antiviral activity and single-dose pharmacokinetics in humans of a novel, orally bioavailable inhibitor of human rhinovirus 3C protease.Antimicrob Agents Chemother. 2005 Jun;49(6):2267-75. doi: 10.1128/AAC.49.6.2267-2275.2005. Antimicrob Agents Chemother. 2005. PMID: 15917520 Free PMC article. Clinical Trial.
-
Proteases of human rhinovirus: role in infection.Methods Mol Biol. 2015;1221:129-41. doi: 10.1007/978-1-4939-1571-2_10. Methods Mol Biol. 2015. PMID: 25261311 Review.
-
Human rhinovirus 3C protease as a potential target for the development of antiviral agents.Curr Protein Pept Sci. 2007 Feb;8(1):19-27. doi: 10.2174/138920307779941523. Curr Protein Pept Sci. 2007. PMID: 17305557 Review.
Cited by
-
In silico molecular screening of bioactive natural compounds of rosemary essential oil and extracts for pharmacological potentials against rhinoviruses.Sci Rep. 2024 Jul 29;14(1):17426. doi: 10.1038/s41598-024-68450-3. Sci Rep. 2024. PMID: 39075176 Free PMC article.
-
Enterovirus infection in Korean children and anti-enteroviral potential candidate agents.Korean J Pediatr. 2012 Oct;55(10):359-66. doi: 10.3345/kjp.2012.55.10.359. Epub 2012 Oct 29. Korean J Pediatr. 2012. PMID: 23133481 Free PMC article.
-
In Vitro Efficacy of Antiviral Compounds against Enterovirus D68.Antimicrob Agents Chemother. 2015 Dec;59(12):7779-81. doi: 10.1128/AAC.00766-15. Epub 2015 Sep 14. Antimicrob Agents Chemother. 2015. PMID: 26149998 Free PMC article.
-
Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.Biochemistry. 2021 Oct 5;60(39):2925-2931. doi: 10.1021/acs.biochem.1c00414. Epub 2021 Sep 10. Biochemistry. 2021. PMID: 34506130 Free PMC article.
-
Antiviral Mechanisms of Saucerneol from Saururus chinensis against Enterovirus A71, Coxsackievirus A16, and Coxsackievirus B3: Role of Mitochondrial ROS and the STING/TKB-1/IRF3 Pathway.Viruses. 2023 Dec 21;16(1):16. doi: 10.3390/v16010016. Viruses. 2023. PMID: 38275951 Free PMC article.
References
-
- Arruda, E., and F. G. Hayden. 1995. Clinical studies of antiviral agents for picornaviral infections, p. 321-355. In D. J. Jeffries and E. De Clerq (ed.), Antiviral chemotherapy. John Wiley & Sons Ltd., Chichester, N.Y.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

