The D4Z4 repeat-mediated pathogenesis of facioscapulohumeral muscular dystrophy
- PMID: 15674778
- PMCID: PMC1196390
- DOI: 10.1086/428361
The D4Z4 repeat-mediated pathogenesis of facioscapulohumeral muscular dystrophy
Figures
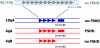
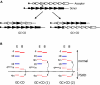
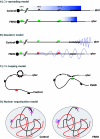
References
Electronic-Database Information
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for FSHD and ICF)
References
-
- Agresti A, Meneveri R, Siccardi AG, Marozzi A, Corneo G, Gaudi S, Ginelli E (1989) Linkage in human heterochromatin between highly divergent Sau3A repeats and a new family of repeated DNA sequences (HaeIII family). J Mol Biol 205:625–631 - PubMed
-
- Bakker E, Wijmenga C, Vossen RH, Padberg GW, Hewitt J, van der Wielen M, Rasmussen K, Frants RR (1995) The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter. Muscle Nerve 2:S39–S44 - PubMed
-
- Ballarati L, Piccini I, Carbone L, Archidiacono N, Rollier A, Marozzi A, Meneveri R, Ginelli E (2002) Human genome dispersal and evolution of 4q35 duplications and interspersed LSau repeats. Gene 296:21–27 - PubMed
-
- Coppee F, Matteotti C, Ansseau E, Sauvage S, Leclercq I, Leroy A, Marcowycz A, Gerbaux C, Figlewicz D, Ding H, Belayew A (2004) The DUX gene family and FSHD. In: Upadhyaya M, Cooper DN (eds) Facioscapulohumeral muscular dystrophy: clinical medicine and molecular cell biology. Garland Science, Oxon, pp 117–134
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

