Imprinting, expression, and localisation of DLK1 in Wilms tumours
- PMID: 15677533
- PMCID: PMC1770562
- DOI: 10.1136/jcp.2004.021717
Imprinting, expression, and localisation of DLK1 in Wilms tumours
Abstract
Background: Loss of imprinting (LOI) of the H19/IGF2 domain is a common feature of Wilms tumour. The GTL2/DLK1 domain is also imprinted and is structurally similar to H19/IGF2. The question arises as to whether DLK1 also undergoes LOI in Wilms tumour, or whether the LOI mechanism is restricted to the H19/IGF2 domain.
Aim: To investigate the imprinting status of DLK1 in Wilms tumours with IGF2 LOI. The cellular localisation of DLK1 in the tumours was also examined.
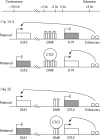
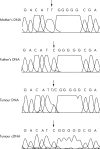
Methods: DLK1 expression was measured by quantitative real time polymerase chain reaction (Q-PCR) in 30 Wilms tumours that had previously been classified according to whether they had IGF2 LOI, WT1 mutations, or 11p15.5 loss of heterozygosity. Allele specific expression of DLK1 was examined by direct sequencing using a DLK1 exon 5 polymorphism (rs1802710). Immunohistochemical analysis of DLK1 was performed on 13 tumours and two intralobar nephrogenic rests, in addition to two fetal kidneys and one fetal skeletal muscle sample.
Results: Ten of 30 tumours were heterozygous for rs1802710 and all tumours showed retention of imprinting of DLK1. Moderate to high expression of DLK1 was detected by Q-PCR in nine of 13 tumours with myogenic differentiation. Immunohistochemical expression of DLK1 was detected in the myogenic elements.
Conclusion: LOI does not occur at the GTL2/DLK1 domain in Wilms tumour. This finding suggests that LOI at 11p15.5 does not reflect non-specific disruption of a shared imprinting mechanism. DLK1 expression in Wilms tumour might reflect the presence of myogenic differentiation, rather than an alteration of its imprinting status.
Figures


Similar articles
-
Epigenetic alteration at the DLK1-GTL2 imprinted domain in human neoplasia: analysis of neuroblastoma, phaeochromocytoma and Wilms' tumour.Br J Cancer. 2005 Apr 25;92(8):1574-80. doi: 10.1038/sj.bjc.6602478. Br J Cancer. 2005. PMID: 15798773 Free PMC article.
-
Wilms tumour histology is determined by distinct types of precursor lesions and not epigenetic changes.J Pathol. 2008 Aug;215(4):377-87. doi: 10.1002/path.2366. J Pathol. 2008. PMID: 18484682
-
Imprinting status of DLK1 gene in brain tumors and lymphomas.Int J Oncol. 2004 Apr;24(4):1011-5. Int J Oncol. 2004. PMID: 15010842
-
Role of genomic imprinting in Wilms' tumour and overgrowth disorders.Med Pediatr Oncol. 1996 Nov;27(5):470-5. doi: 10.1002/(SICI)1096-911X(199611)27:5<470::AID-MPO14>3.0.CO;2-E. Med Pediatr Oncol. 1996. PMID: 8827076 Review.
-
Genomic imprinting and carcinogenesis.Histol Histopathol. 1998 Apr;13(2):425-35. doi: 10.14670/HH-13.425. Histol Histopathol. 1998. PMID: 9589901 Review.
Cited by
-
Abnormally localized DLK1 interacts with NCOR1 in non-small cell lung cancer cell nuclear.Biosci Rep. 2019 Dec 20;39(12):BSR20192362. doi: 10.1042/BSR20192362. Biosci Rep. 2019. PMID: 31661545 Free PMC article.
-
The regulation of non-coding RNA expression in the liver of mice fed DDC.Exp Mol Pathol. 2009 Aug;87(1):12-9. doi: 10.1016/j.yexmp.2009.03.006. Epub 2009 Apr 9. Exp Mol Pathol. 2009. PMID: 19362547 Free PMC article.
-
Analysis of the Paternally-Imprinted DLK1-MEG3 and IGF2-H19 Tandem Gene Loci in NT2 Embryonal Carcinoma Cells Identifies DLK1 as a Potential Therapeutic Target.Stem Cell Rev Rep. 2018 Dec;14(6):823-836. doi: 10.1007/s12015-018-9838-5. Stem Cell Rev Rep. 2018. PMID: 29980981
-
Genes of DLK1-DIO3 Locus and miR-379/656 Cluster is a Potential Diagnostic and Prognostic Marker in Patients With Hepatocellular Carcinoma: A Systems Biology Study.J Clin Exp Hepatol. 2025 Mar-Apr;15(2):102450. doi: 10.1016/j.jceh.2024.102450. Epub 2024 Nov 12. J Clin Exp Hepatol. 2025. PMID: 39698049
-
Emerging Roles of DLK1 in the Stem Cell Niche and Cancer Stemness.J Histochem Cytochem. 2022 Jan;70(1):17-28. doi: 10.1369/00221554211048951. Epub 2021 Oct 4. J Histochem Cytochem. 2022. PMID: 34606325 Free PMC article. Review.
References
-
- Zemel S, Bartolomei MS, Tilghman SM. Physical linkage of two mammalian imprinted genes, H19 and insulin-like growth factor 2. Nat Genet 1992;2:61–5. - PubMed
-
- Leighton PA, Saam JR, Ingram RS, et al. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev 1995;9:2079–89. - PubMed
-
- Frevel MA, Sowerby SJ, Petersen GB, et al. Methylation sequencing analysis refines the region of H19 epimutation in Wilms tumor. J Biol Chem 1999;274:29331–40. - PubMed
-
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 2000;405:482–5. - PubMed
-
- Hark AT, Schoenherr CJ, Katz DJ, et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 2000;405:486–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
