Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients
- PMID: 15677557
- PMCID: PMC1895072
- DOI: 10.1182/blood-2004-09-3704
Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients
Abstract
Multiple myeloma (MM) is a B-lineage malignancy characterized by diverse genetic subtypes and clinical outcomes. The recurrent immunoglobulin heavy chain (IgH) switch translocation, t(4;14)(p16;q32), is associated with poor outcome, though the mechanism is unclear. Quantitative reverse-transcription-polymerase chain reaction (RT-PCR) for proposed target genes on a panel of myeloma cell lines and purified plasma cells showed that only transcripts originating from the WHSC1/MMSET/NSD2 gene are uniformly dysregulated in all t(4;14)POS patients. The different transcripts detected, multiple myeloma SET domain containing protein (MMSET I), MMSET II, Exon 4a/MMSET III, and response element II binding protein (RE-IIBP), are produced by alternative splicing and alternative transcription initiation events. Translation of the various transcripts, including those from major breakpoint region 4-2 (MB4-2) and MB4-3 breakpoint variants, was confirmed by transient transfection and immunoblotting. Green fluorescent protein (GFP)-tagged MMSET I and II, corresponding to proteins expressed in MB4-1 patients, localized to the nucleus but not nucleoli, whereas the MB4-2 and MB4-3 proteins concentrate in nucleoli. Cloning and localization of the Exon 4a/MMSET III splice variant, which contains the protein segment lost in the MB4-2 variant, identified a novel protein domain that prevents nucleolar localization. Kinetic studies using photobleaching suggest that the breakpoint variants are functionally distinct from wild-type proteins. In contrast, RE-IIBP is universally dysregulated and also potentially functional in all t(4;14)POS patients irrespective of fibroblast growth factor receptor 3 (FGFR3) expression or breakpoint type.
Figures

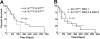
 represents t(4;14)POS FGFR3 nonexpressing patients. Patients, t(4;14)POS, were selected on the basis of a BM plasmacytosis more than 35%, and then best match t(4;14)NEG samples were selected based on sex, M-protein isotype, bone marrow plasmacytosis, and age. Error bars represent the standard deviation.
represents t(4;14)POS FGFR3 nonexpressing patients. Patients, t(4;14)POS, were selected on the basis of a BM plasmacytosis more than 35%, and then best match t(4;14)NEG samples were selected based on sex, M-protein isotype, bone marrow plasmacytosis, and age. Error bars represent the standard deviation.



Similar articles
-
The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts.Blood. 1998 Nov 1;92(9):3025-34. Blood. 1998. PMID: 9787135
-
Identification of ID-1 as a potential target gene of MMSET in multiple myeloma.Br J Haematol. 2005 Sep;130(5):700-8. doi: 10.1111/j.1365-2141.2005.05664.x. Br J Haematol. 2005. PMID: 16115125
-
A molecular study of the t(4;14) in multiple myeloma.Br J Haematol. 2002 Aug;118(2):514-20. doi: 10.1046/j.1365-2141.2002.03618.x. Br J Haematol. 2002. PMID: 12139740
-
MMSET: role and therapeutic opportunities in multiple myeloma.Biomed Res Int. 2014;2014:636514. doi: 10.1155/2014/636514. Epub 2014 Jul 1. Biomed Res Int. 2014. PMID: 25093175 Free PMC article. Review.
-
Ten years and counting: so what do we know about t(4;14)(p16;q32) multiple myeloma.Leuk Lymphoma. 2006 Nov;47(11):2289-300. doi: 10.1080/10428190600822128. Leuk Lymphoma. 2006. PMID: 17107900 Review.
Cited by
-
MMSET regulates expression of IRF4 in t(4;14) myeloma and its silencing potentiates the effect of bortezomib.Leukemia. 2015 Dec;29(12):2347-54. doi: 10.1038/leu.2015.169. Epub 2015 Jun 30. Leukemia. 2015. PMID: 26196464
-
Gene Fusions Create Partner and Collateral Dependencies Essential to Cancer Cell Survival.Cancer Res. 2021 Aug 1;81(15):3971-3984. doi: 10.1158/0008-5472.CAN-21-0791. Epub 2021 Jun 7. Cancer Res. 2021. PMID: 34099491 Free PMC article.
-
NSD2 promotes tumor angiogenesis through methylating and activating STAT3 protein.Oncogene. 2021 Apr;40(16):2952-2967. doi: 10.1038/s41388-021-01747-z. Epub 2021 Mar 19. Oncogene. 2021. PMID: 33742125
-
Plasma membrane proteomics identifies biomarkers associated with MMSET overexpression in T(4;14) multiple myeloma.Oncotarget. 2013 Jul;4(7):1008-18. doi: 10.18632/oncotarget.1049. Oncotarget. 2013. PMID: 23900284 Free PMC article.
-
WHSC1 acts as a prognostic indicator and functions as an oncogene in cervical cancer.Onco Targets Ther. 2019 Jun 17;12:4683-4690. doi: 10.2147/OTT.S204701. eCollection 2019. Onco Targets Ther. 2019. PMID: 31354300 Free PMC article.
References
-
- Avet-Loiseau H, Facon T, Grosbois B, et al. Oncogenesis of multiple myeloma: 14q32 and 13q chromosomal abnormalities are not randomly distributed, but correlate with natural history, immunological features, and clinical presentation. Blood. 2002;99: 2185-2191. - PubMed
-
- Moreau P, Facon T, Leleu X, et al. Recurrent 14q32 translocations determine the prognosis of multiple myeloma, especially in patients receiving intensive chemotherapy. Blood. 2002;100: 1579-1583. - PubMed
-
- Nishida K, Tamura A, Nakazawa N, et al. The Ig heavy chain gene is frequently involved in chromosomal translocations in multiple myeloma and plasma cell leukemia as detected by in situ hybridization. Blood. 1997;90: 526-534. - PubMed
-
- Fenton JA, Pratt G, Rawstron AC, et al. Genomic characterization of the chromosomal breakpoints of t(4;14) of multiple myeloma suggests more than one possible aetiological mechanism. Oncogene. 2003;22: 1103-1113. - PubMed
-
- Fenton JA, Pratt G, Rothwell DG, Rawstron AC, Morgan GJ. Translocation t(11;14) in multiple myeloma: analysis of translocation breakpoints on der(11) and der(14) chromosomes suggests complex molecular mechanisms of recombination. Genes Chromosomes Cancer. 2004;39: 151-155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

