NMR screening and crystal quality of bacterially expressed prokaryotic and eukaryotic proteins in a structural genomics pipeline
- PMID: 15677718
- PMCID: PMC548552
- DOI: 10.1073/pnas.0408490102
NMR screening and crystal quality of bacterially expressed prokaryotic and eukaryotic proteins in a structural genomics pipeline
Abstract
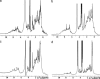
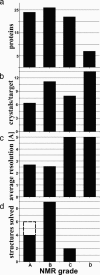
In the Joint Center for Structural Genomics, one-dimensional (1D) 1H NMR spectroscopy is routinely used to characterize the folded state of protein targets and, thus, serves to guide subsequent crystallization efforts and to identify proteins for NMR structure determination. Here, we describe 1D 1H NMR screening of a group of 79 mouse homologue proteins, which correlates the NMR data with the outcome of subsequent crystallization experiments and crystallographic structure determination. Based on the 1D 1H NMR spectra, the proteins are classified into four groups, "A" to "D." A-type proteins are candidates for structure determination by NMR or crystallography; "B"-type are earmarked for crystallography; "C" indicates folded globular proteins with broadened line shapes; and "D" are nonglobular, "unfolded" polypeptides. The results obtained from coarse- and fine-screen crystallization trials imply that only A- and B-type proteins should be used for extensive crystallization trials in the future, with C and D proteins subjected only to coarse-screen crystallization trials. Of the presently studied 79 soluble protein targets, 63% yielded A- or B-quality 1D 1H NMR spectra. Although similar yields of crystallization hits were obtained for all four groups, A to D, crystals from A- and B-type proteins diffracted on average to significantly higher resolution than crystals produced from C- or D-type proteins. Furthermore, the output of refined crystal structures from this test set of proteins was 4-fold higher for A- and B-type than for C- and D-type proteins.
Figures

Similar articles
-
Towards miniaturization of a structural genomics pipeline using micro-expression and microcoil NMR.J Struct Funct Genomics. 2005 Dec;6(4):259-67. doi: 10.1007/s10969-005-9000-x. Epub 2005 Nov 9. J Struct Funct Genomics. 2005. PMID: 16283429
-
Strategies for improving crystallization success rates.Methods Mol Biol. 2008;426:345-62. doi: 10.1007/978-1-60327-058-8_22. Methods Mol Biol. 2008. PMID: 18542875
-
Comparisons of NMR spectral quality and success in crystallization demonstrate that NMR and X-ray crystallography are complementary methods for small protein structure determination.J Am Chem Soc. 2005 Nov 30;127(47):16505-11. doi: 10.1021/ja053564h. J Am Chem Soc. 2005. PMID: 16305237
-
Protein crystallization: virtual screening and optimization.Prog Biophys Mol Biol. 2005 Jul;88(3):285-309. doi: 10.1016/j.pbiomolbio.2004.07.008. Epub 2004 Sep 30. Prog Biophys Mol Biol. 2005. PMID: 15652246 Review.
-
A community resource of experimental data for NMR / X-ray crystal structure pairs.Protein Sci. 2016 Jan;25(1):30-45. doi: 10.1002/pro.2774. Epub 2015 Sep 22. Protein Sci. 2016. PMID: 26293815 Free PMC article. Review.
Cited by
-
Structural basis for a Munc13-1 homodimer to Munc13-1/RIM heterodimer switch.PLoS Biol. 2006 Jul;4(7):e192. doi: 10.1371/journal.pbio.0040192. PLoS Biol. 2006. PMID: 16732694 Free PMC article.
-
Combinatorial Domain Hunting: An effective approach for the identification of soluble protein domains adaptable to high-throughput applications.Protein Sci. 2006 Oct;15(10):2356-65. doi: 10.1110/ps.062082606. Protein Sci. 2006. PMID: 17008718 Free PMC article.
-
Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7.J Virol. 2005 Oct;79(20):12905-13. doi: 10.1128/JVI.79.20.12905-12913.2005. J Virol. 2005. PMID: 16188992 Free PMC article.
-
The boxing glove shape of subunit d of the yeast V-ATPase in solution and the importance of disulfide formation for folding of this protein.J Bioenerg Biomembr. 2007 Aug;39(4):275-89. doi: 10.1007/s10863-007-9089-7. Epub 2007 Sep 26. J Bioenerg Biomembr. 2007. PMID: 17896169
-
High-throughput screening of optimal solution conditions for structural biological studies by fluorescence correlation spectroscopy.Protein Sci. 2009 May;18(5):1115-20. doi: 10.1002/pro.92. Protein Sci. 2009. PMID: 19388076 Free PMC article.
References
-
- McDonald, C. C. & Phillips, W. D. (1967) J. Am. Chem. Soc. 89, 6332–6341. - PubMed
-
- Edwards, A. M., Arrowsmith, C. H., Christendat, D., Dharamsi, A., Friesen, J. D., Greenblatt, J. F. & Vedadi, M. (2000) Nat. Struct. Biol. 7, Suppl., 970–972. - PubMed
-
- Prestegard, J. H., Valafar, H., Glushka, J. & Tian, F. (2001) Biochemistry 40, 8677–8685. - PubMed
-
- Rehm, T., Huber, R. & Holak, T. A. (2002) Structure (London) 10, 1613–1618. - PubMed
-
- Kennedy, M. A., Montelione, G. T., Arrowsmith, C. H. & Markley, J. L. (2002) J. Struct. Funct. Genomics 2, 155–169. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

