CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator
- PMID: 15678159
- PMCID: PMC1299244
- DOI: 10.1038/sj.embor.7400334
CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator
Abstract
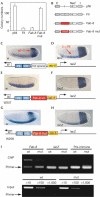
Eukaryotic transcriptional regulation often involves regulatory elements separated from the cognate genes by long distances, whereas appropriately positioned insulator or enhancer-blocking elements shield promoters from illegitimate enhancer action. Four proteins have been identified in Drosophila mediating enhancer blocking-Su(Hw), Zw5, BEAF32 and GAGA factor. In vertebrates, the single protein CTCF, with 11 highly conserved zinc fingers, confers enhancer blocking in all known chromatin insulators. Here, we characterize an orthologous CTCF factor in Drosophila with a similar domain structure, binding site specificity and transcriptional repression activity as in vertebrates. In addition, we demonstrate that one of the insulators (Fab-8) in the Drosophila Abdominal-B locus mediates enhancer blocking by dCTCF. Therefore, the enhancer-blocking protein CTCF and, most probably, the mechanism of enhancer blocking mediated by this remarkably versatile factor are conserved from Drosophila to humans.
Figures



References
-
- Baniahmad A, Steiner C, Kohne AC, Renkawitz R (1990) Modular structure of a chicken lysozyme silencer: involvement of an unusual thyroid hormone receptor binding site. Cell 61: 505–514 - PubMed
-
- Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, Schedl P, Gyurkovics H, Karch F (2000) The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127: 779–790 - PubMed
-
- Bell AC, West AG, Felsenfeld G (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98: 387–396 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

