Programmed ribosomal frameshifting in decoding the SARS-CoV genome
- PMID: 15680415
- PMCID: PMC7111862
- DOI: 10.1016/j.virol.2004.11.038
Programmed ribosomal frameshifting in decoding the SARS-CoV genome
Abstract
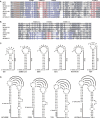
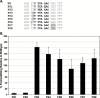
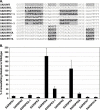
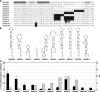
Programmed ribosomal frameshifting is an essential mechanism used for the expression of orf1b in coronaviruses. Comparative analysis of the frameshift region reveals a universal shift site U_UUA_AAC, followed by a predicted downstream RNA structure in the form of either a pseudoknot or kissing stem loops. Frameshifting in SARS-CoV has been characterized in cultured mammalian cells using a dual luciferase reporter system and mass spectrometry. Mutagenic analysis of the SARS-CoV shift site and mass spectrometry of an affinity tagged frameshift product confirmed tandem tRNA slippage on the sequence U_UUA_AAC. Analysis of the downstream pseudoknot stimulator of frameshifting in SARS-CoV shows that a proposed RNA secondary structure in loop II and two unpaired nucleotides at the stem I-stem II junction in SARS-CoV are important for frameshift stimulation. These results demonstrate key sequences required for efficient frameshifting, and the utility of mass spectrometry to study ribosomal frameshifting.
Figures

References
-
- Baranov P.V., Gesteland R.F., Atkins J.F. Recoding: translational bifurcations in gene expression. Gene. 2002;286(2):187–201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

