RNA recombination of hepatitis delta virus in natural mixed-genotype infection and transfected cultured cells
- PMID: 15681424
- PMCID: PMC546541
- DOI: 10.1128/JVI.79.4.2221-2229.2005
RNA recombination of hepatitis delta virus in natural mixed-genotype infection and transfected cultured cells
Abstract
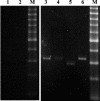
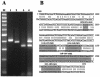
Most RNA viruses encode their own RNA polymerases for genome replication, and increasing numbers of them appear to be capable of undergoing RNA recombination. Here, we provide the first report of intergenotypic recombination in hepatitis delta virus (HDV), the only animal RNA virus that requires host RNA polymerase(s) for viral replication. In vivo, we analyzed RNA species derived from the serum of a patient with mixed genotype I and genotype IIb HDV infection by using multiple restriction fragment length polymorphism (RFLP) assays and sequence analysis of cloned reverse transcription (RT)-PCR products. Six HDV recombinants were isolated from 101 tested clones, and HDV recombination in this patient was further confirmed by RT-PCR with genotype-specific primer pairs. Analysis of the recombination junctions suggested that the HDV genome rearrangement occurred through faithful homologous recombination. We then used an RNA cotransfection cell culture system to investigate HDV RNA recombination in vitro. We found that HDV recombinants could indeed be detected in the transfected cells; some of these possessed recombination junctions identical to those identified in vivo. Furthermore, using a PCR-independent RNase protection assay, we were able to readily identify the recombined HDV RNA species in cultured cells. Taken together, our results demonstrate that HDV RNA recombination occurs in both natural HDV infections and cultured cells, thereby presenting a novel mechanism for HDV evolution.
Figures




References
-
- Ambros, S., J. C. Desvignes, G. Llacer, and R. Flores. 1995. Pear blister canker viroid: sequence variability and causal role in pear blister canker disease. J. Gen. Virol. 76:2625-2629. - PubMed
-
- Branch, A. D., B. J. Benenfeld, B. M. Baroudy, F. V. Wells, J. L. Gerin, and H. D. Robertson. 1989. An ultraviolet-sensitive RNA structural element in a viroid-like domain of the hepatitis delta virus. Science 243:649-652. - PubMed
-
- Brazas, R., and D. Ganem. 1996. A cellular homolog of hepatitis delta antigen: implications for viral replication and evolution. Science 274:90-94. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

