The topology of bulges in the long stem of the flavivirus 3' stem-loop is a major determinant of RNA replication competence
- PMID: 15681432
- PMCID: PMC546603
- DOI: 10.1128/JVI.79.4.2309-2324.2005
The topology of bulges in the long stem of the flavivirus 3' stem-loop is a major determinant of RNA replication competence
Abstract
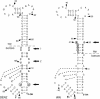
All flavivirus genomes contain a 3'terminal stem-loop secondary structure (3'SL) formed by the most downstream approximately 100 nucleotides (nt) of the viral RNA. The 3'SL is required for virus replication and has been shown to bind both virus-coded and cellular proteins. Results of the present study using an infectious DNA for WN virus strain 956 initially demonstrated that the dengue virus serotype 2 (DEN2) 3'SL nucleotide sequence could not substitute for that of the WN 3'SL to support WN genome replication. To determine what WN virus-specific 3'SL nucleotide sequences were required for WN virus replication, WN virus 3'SL nucleotide sequences were selectively deleted and replaced by analogous segments of the DEN2 3'SL nucleotide sequence such that the overall 3'SL secondary structure was not disrupted. Top and bottom portions of the WN virus 3'SL were defined according to previous studies (J. L. Blackwell and M. A. Brinton, J. Virol. 71:6433-6444, 1997; L. Zeng, L., B. Falgout, and L. Markoff, J. Virol. 72:7510-7522, 1998). A bulge in the top portion of the long stem of the WN 3'SL was essential for replication of mutant WN RNAs, and replication-defective RNAs failed to produce negative strands in transfected cells. Introduction of a second bulge into the bottom portion of the long stem of the wild-type WN 3'SL markedly enhanced the replication competence of WN virus in mosquito cells but had no effect on replication in mammalian cells. This second bulge was identified as a host cell-specific enhancer of flavivirus replication. Results suggested that bulges and their topological location within the long stem of the 3'SL are primary determinants of replication competence for flavivirus genomes.
Figures




Similar articles
-
An RNA Thermometer Activity of the West Nile Virus Genomic 3'-Terminal Stem-Loop Element Modulates Viral Replication Efficiency during Host Switching.Viruses. 2020 Jan 15;12(1):104. doi: 10.3390/v12010104. Viruses. 2020. PMID: 31952291 Free PMC article.
-
Enhancement of dengue virus translation: role of the 3' untranslated region and the terminal 3' stem-loop domain.Virology. 2004 Nov 10;329(1):119-33. doi: 10.1016/j.virol.2004.08.004. Virology. 2004. PMID: 15476880
-
BHK cell proteins that bind to the 3' stem-loop structure of the West Nile virus genome RNA.J Virol. 1995 Sep;69(9):5650-8. doi: 10.1128/JVI.69.9.5650-5658.1995. J Virol. 1995. PMID: 7637011 Free PMC article.
-
The structure-function relationship of the enterovirus 3'-UTR.Virus Res. 2009 Feb;139(2):209-16. doi: 10.1016/j.virusres.2008.07.014. Epub 2008 Aug 30. Virus Res. 2009. PMID: 18706945 Review.
-
[Structure and function of the non-coding regions of hepatitis C viral RNA].Postepy Biochem. 2006;52(1):62-71. Postepy Biochem. 2006. PMID: 16869303 Review. Polish.
Cited by
-
Functional analysis of the tick-borne encephalitis virus cyclization elements indicates major differences between mosquito-borne and tick-borne flaviviruses.J Virol. 2006 Apr;80(8):4099-113. doi: 10.1128/JVI.80.8.4099-4113.2006. J Virol. 2006. PMID: 16571826 Free PMC article.
-
A 5' RNA element promotes dengue virus RNA synthesis on a circular genome.Genes Dev. 2006 Aug 15;20(16):2238-49. doi: 10.1101/gad.1444206. Epub 2006 Aug 1. Genes Dev. 2006. PMID: 16882970 Free PMC article.
-
In Silico Analysis of Dengue Virus Serotype 2 Mutations Detected at the Intrahost Level in Patients with Different Clinical Outcomes.Microbiol Spectr. 2021 Oct 31;9(2):e0025621. doi: 10.1128/Spectrum.00256-21. Epub 2021 Sep 1. Microbiol Spectr. 2021. PMID: 34468189 Free PMC article.
-
Genomic and phylogenetic characterization of Brazilian yellow fever virus strains.J Virol. 2012 Dec;86(24):13263-71. doi: 10.1128/JVI.00565-12. Epub 2012 Sep 26. J Virol. 2012. PMID: 23015713 Free PMC article.
-
Zika virus: An emerging flavivirus.J Microbiol. 2017 Mar;55(3):204-219. doi: 10.1007/s12275-017-7063-6. Epub 2017 Feb 28. J Microbiol. 2017. PMID: 28243937 Review.
References
-
- Bartel, D. P., M. L. Zapp, M. R. Green, and J. W. Szostak. 1991. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in the viral RNA. Cell 17:529-536. - PubMed
-
- Beasley, D. L., L. Li, M. T. Suderman, and A. D. Barrett. 2002. Mouse neuroinvasiveness phenotype of West Nile virus strains varies depending upon virus genotype. Virology 296:17-23. - PubMed
-
- Brinton, M. A., A. V. Fernandez, and J. H. Dispoto. 1986. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology 153:113-121. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

