Analysis of the interaction of the adenovirus L1 52/55-kilodalton and IVa2 proteins with the packaging sequence in vivo and in vitro
- PMID: 15681437
- PMCID: PMC546600
- DOI: 10.1128/JVI.79.4.2366-2374.2005
Analysis of the interaction of the adenovirus L1 52/55-kilodalton and IVa2 proteins with the packaging sequence in vivo and in vitro
Erratum in
- J Virol. 2006 Feb;80(3):1614
Abstract
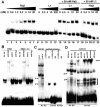
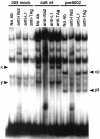
We previously showed that the adenovirus IVa2 and L1 52/55-kDa proteins interact in infected cells and the IVa2 protein is part of two virus-specific complexes (x and y) formed in vitro with repeated elements of the packaging sequence called the A1-A2 repeats. Here we demonstrate that both the IVa2 and L1 52/55-kDa proteins bind in vivo to the packaging sequence and that each protein-DNA interaction is independent of the other. There is a strong and direct interaction of the IVa2 protein with DNA in vitro. This interaction is observed when probes containing the A1-A2 or A4-A5 repeats are used, but it is not found by using an A5-A6 probe. Furthermore, we show that complex x is likely a heterodimer of IVa2 and an unknown viral protein, while complex y is a monomer or multimer of IVa2. No in vitro interaction of purified L1 52/55-kDa protein with the packaging sequence was found, suggesting that the L1 52/55-kDa protein-DNA interaction may be mediated by an intermediate protein. Results support roles for both the L1 52/55-kDa and IVa2 proteins in DNA encapsidation.
Figures



Similar articles
-
Dependence of the encapsidation function of the adenovirus L1 52/55-kilodalton protein on its ability to bind the packaging sequence.J Virol. 2006 Feb;80(4):1965-71. doi: 10.1128/JVI.80.4.1965-1971.2006. J Virol. 2006. PMID: 16439552 Free PMC article.
-
Functional interaction of the adenovirus IVa2 protein with adenovirus type 5 packaging sequences.J Virol. 2005 Mar;79(5):2831-8. doi: 10.1128/JVI.79.5.2831-2838.2005. J Virol. 2005. PMID: 15709002 Free PMC article.
-
Interaction of the adenovirus major core protein precursor, pVII, with the viral DNA packaging machinery.Virology. 2005 Apr 10;334(2):194-202. doi: 10.1016/j.virol.2005.01.048. Virology. 2005. PMID: 15780869
-
Requirement of the adenovirus IVa2 protein for virus assembly.J Virol. 2003 Mar;77(6):3586-94. doi: 10.1128/jvi.77.6.3586-3594.2003. J Virol. 2003. PMID: 12610134 Free PMC article.
-
Interaction of the adenovirus IVa2 protein with viral packaging sequences.J Virol. 2000 Mar;74(6):2687-93. doi: 10.1128/jvi.74.6.2687-2693.2000. J Virol. 2000. PMID: 10684284 Free PMC article.
Cited by
-
Role for the L1-52/55K protein in the serotype specificity of adenovirus DNA packaging.J Virol. 2008 May;82(10):5089-92. doi: 10.1128/JVI.00040-08. Epub 2008 Mar 12. J Virol. 2008. PMID: 18337584 Free PMC article.
-
Dependence of the encapsidation function of the adenovirus L1 52/55-kilodalton protein on its ability to bind the packaging sequence.J Virol. 2006 Feb;80(4):1965-71. doi: 10.1128/JVI.80.4.1965-1971.2006. J Virol. 2006. PMID: 16439552 Free PMC article.
-
Ternary complex formation on the adenovirus packaging sequence by the IVa2 and L4 22-kilodalton proteins.J Virol. 2007 Nov;81(22):12450-7. doi: 10.1128/JVI.01470-07. Epub 2007 Sep 5. J Virol. 2007. PMID: 17804492 Free PMC article.
-
Adenovirus structural protein IIIa is involved in the serotype specificity of viral DNA packaging.J Virol. 2011 Aug;85(15):7849-55. doi: 10.1128/JVI.00467-11. Epub 2011 Jun 1. J Virol. 2011. PMID: 21632753 Free PMC article.
-
The adenovirus L4 33-kilodalton protein binds to intragenic sequences of the major late promoter required for late phase-specific stimulation of transcription.J Virol. 2007 Feb;81(3):1327-38. doi: 10.1128/JVI.01584-06. Epub 2006 Nov 8. J Virol. 2007. PMID: 17093188 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

