Norovirus proteinase-polymerase and polymerase are both active forms of RNA-dependent RNA polymerase
- PMID: 15681440
- PMCID: PMC546540
- DOI: 10.1128/JVI.79.4.2393-2403.2005
Norovirus proteinase-polymerase and polymerase are both active forms of RNA-dependent RNA polymerase
Abstract
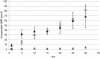
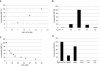
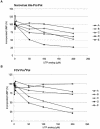
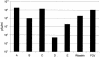
In vitro mapping studies of the MD145 norovirus (Caliciviridae) ORF1 polyprotein identified two stable cleavage products containing the viral RNA-dependent RNA polymerase (RdRp) domains: ProPol (a precursor comprised of both the proteinase and polymerase) and Pol (the mature polymerase). The goal of this study was to identify the active form (or forms) of the norovirus polymerase. The recombinant ProPol (expressed as Pro(-)Pol with an inactivated proteinase domain to prevent autocleavage) and recombinant Pol were purified after synthesis in bacteria and shown to be active RdRp enzymes. In addition, the mutant His-E1189A-ProPol protein (with active proteinase but with the natural ProPol cleavage site blocked) was active as an RdRp, confirming that the norovirus ProPol precursor could possess two enzymatic activities simultaneously. The effects of several UTP analogs on the RdRp activity of the norovirus and feline calicivirus Pro(-)Pol enzymes were compared and found to be similar. Our data suggest that the norovirus ProPol is a bifunctional enzyme during virus replication. The availability of this recombinant ProPol enzyme might prove useful in the development of antiviral drugs for control of the noroviruses associated with acute gastroenteritis.
Figures




References
-
- Arnold, J. J., and C. E. Cameron. 1999. Poliovirus RNA-dependent RNA polymerase (3Dpol) is sufficient for template switching in vitro. J. Biol. Chem. 274:2706-2716. - PubMed
-
- Belliot, G., S. V. Sosnovtsev, T. Mitra, C. Hammer, M. Garfield, and K. Y. Green. 2003. In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells. J. Virol. 77:10957-10974. - PMC - PubMed
-
- Blakeney, S. J., A. Cahill, and P. A. Reilly. 2003. Processing of Norwalk virus nonstructural proteins by a 3C-like cysteine proteinase. Virology 308:216-224. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

