A multi-exonic BRCA1 deletion identified in multiple families through single nucleotide polymorphism haplotype pair analysis and gene amplification with widely dispersed primer sets
- PMID: 15681486
- PMCID: PMC1867506
- DOI: 10.1016/S1525-1578(10)60020-7
A multi-exonic BRCA1 deletion identified in multiple families through single nucleotide polymorphism haplotype pair analysis and gene amplification with widely dispersed primer sets
Abstract
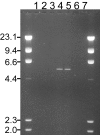
The identification of intragenic rearrangements is important for a comprehensive understanding of mutations that occur in some clinically important genes. Single nucleotide polymorphism haplotypes obtained from clinical sequence data have been used to identify patients at high risk for rearrangement mutations. Application of this method identified a novel 26-kb deletion of BRCA1 exons 14 through 20 in patients from multiple families with hereditary breast and ovarian cancer. Clinical sequence data from 5911 anonymous patients were screened for genotypes that were inconsistent with known pairs of canonical haplotypes in BRCA1 that could be explained by hemizygous deletions involving exon 16. Long-range polymerase chain reaction demonstrated that two of six samples identified by this search contained a deletion in the expected region encompassing exons 14 through 20. The breakpoint was fully characterized by DNA sequencing and demonstrated that the deletion resulted from Alu-mediated recombination. This mutation was also identified twice in a set of 982 anonymous specimens that had negative clinical test results, but uninformative haplotypes. Three additional occurrences of this mutation were found by testing 10 other patients with the indicative genotype. An assay for this mutation was added to a comprehensive clinical breast/ovarian cancer test and eight more instances were found in 20,649 probands. This multiexon deletion has therefore been detected in 15 different North American families with hereditary breast/ovarian cancer. In conclusion, this primarily computational approach is highly effective and identifies specimens using existing data that are enriched for deletion mutations.
Figures
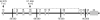

Similar articles
-
Screening for genomic rearrangements in BRCA1 and BRCA2 genes in Czech high-risk breast/ovarian cancer patients: high proportion of population specific alterations in BRCA1 gene.Breast Cancer Res Treat. 2010 Nov;124(2):337-47. doi: 10.1007/s10549-010-0745-y. Epub 2010 Feb 5. Breast Cancer Res Treat. 2010. PMID: 20135348
-
Identification of a 3 kb Alu-mediated BRCA1 gene rearrangement in two breast/ovarian cancer families.Oncogene. 1999 Jul 15;18(28):4160-5. doi: 10.1038/sj.onc.1202754. Oncogene. 1999. PMID: 10435598
-
Identification of a novel BRCA1 large genomic rearrangement in a Spanish breast/ovarian cancer family.Breast Cancer Res Treat. 2008 Nov;112(1):63-7. doi: 10.1007/s10549-007-9839-6. Epub 2007 Dec 1. Breast Cancer Res Treat. 2008. PMID: 18060491
-
Mutation analysis and characterization of ATR sequence variants in breast cancer cases from high-risk French Canadian breast/ovarian cancer families.BMC Cancer. 2006 Sep 29;6:230. doi: 10.1186/1471-2407-6-230. BMC Cancer. 2006. PMID: 17010193 Free PMC article.
-
Characterization of four novel BRCA2 large genomic rearrangements in Spanish breast/ovarian cancer families: review of the literature, and reevaluation of the genetic mechanisms involved in their origin.Breast Cancer Res Treat. 2012 May;133(1):273-83. doi: 10.1007/s10549-011-1909-0. Epub 2012 Mar 21. Breast Cancer Res Treat. 2012. PMID: 22434521 Review.
Cited by
-
Homozygous deletion of the STK11/LKB1 locus and the generation of novel fusion transcripts in cervical cancer cells.Cancer Genet Cytogenet. 2010 Mar;197(2):130-41. doi: 10.1016/j.cancergencyto.2009.11.017. Cancer Genet Cytogenet. 2010. PMID: 20193846 Free PMC article.
-
The relative contribution of point mutations and genomic rearrangements in BRCA1 and BRCA2 in high-risk breast cancer families.Cancer Res. 2008 Sep 1;68(17):7006-14. doi: 10.1158/0008-5472.CAN-08-0599. Epub 2008 Aug 14. Cancer Res. 2008. PMID: 18703817 Free PMC article.
-
Cross-sectional analysis of germline BRCA1 and BRCA2 mutations in Japanese patients suspected to have hereditary breast/ovarian cancer.Cancer Sci. 2008 Oct;99(10):1967-76. doi: 10.1111/j.1349-7006.2008.00944.x. Cancer Sci. 2008. PMID: 19016756 Free PMC article.
References
-
- Frank TS, Deffenbaugh AM, Reid JE, Hulick M, Ward BE, Lingenfelter B, Gumpper KL, Scholl T, Tavtigian SV, Pruss DR, Critchfield GC. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20:1480–1490. - PubMed
-
- Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, Weber B, Lenoir G, Chang-Claude J, Sobol H, Teare MD, Struewing J, Arason A, Scherneck S, Peto J, Rebbeck TR, Tonin P, Neuhausen S, Barkardottir R, Eyfjord J, Lynch H, Ponder BAJ, Gayther SA, Birch JM, Lindblom A, Stoppa-Lyonnet D, Bignon Y, Borg A, Hamann U, Haites N, Scott RJ, Maugard CM, Vasen H, Seitz S, Cannon-Albright LA, Schofield A, Zelada-Hedman M, Breast Cancer Linkage Consortium Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am J Hum Genet. 1998;62:676–689. - PMC - PubMed
-
- Hendrickson BC, Deffenbaugh AM, Gaglio CA, Judkins T, Chen S, Ward BE, Scholl T. Prevalence results for five recurrent BRCA1 rearrangement mutations in 7570 analyses. Am J Hum Genet. 2003;73:253.
-
- Petri-Bosch A, Peelen T, van Vliet M, van Eijk R, Olmer R, Drusedau M, Hogervorst FBL, Hageman S, Arts PJW, Ligtenberg MJL, Meijers-Heijboer H, Klijn JGM, Vasen HFA, Cornelisse CJ, van’t Veer LJ, Bakker E, van Ommen G-JB, Devilee P. BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients. Nat Genet. 1997;17:341–345. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

