Proton pathways in green fluorescence protein
- PMID: 15681647
- PMCID: PMC1305344
- DOI: 10.1529/biophysj.104.055541
Proton pathways in green fluorescence protein
Abstract
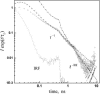
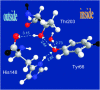
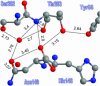
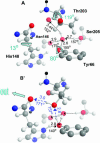
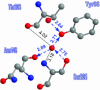
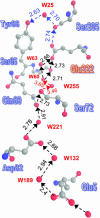
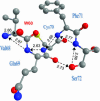
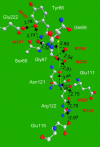
Proton pathways in green fluorescent protein (GFP) are more extended than previously reported. In the x-ray data of wild-type GFP, a two-step exit pathway exists from the active site to the protein surface, controlled by a threonine switch. A proton entry pathway begins at a glutamate-lysine cluster around Glu-5, and extends all the way to the buried Glu-222 near the active site. This structural evidence suggests that GFP may function as a portable light-driven proton-pump, with proton emitted in the excited state through the switchable exit pathway, and replenished from Glu-222 and the Glu-5 entry pathway in the ground state.
Figures

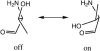


References
-
- Ädelroth, P., and P. Brzezinski. 2004. Surface-mediated proton-transfer reactions in membrane-bound proteins. Biochim. Biophys. Acta. 1655:102–115. - PubMed
-
- Agmon, N. 1995. The Grotthuss mechanism. Chem. Phys. Lett. 244:456–462.
-
- Agmon, N. 2005. Elementary steps in excited-state proton transfer. J. Phys. Chem. A. 109:13–35. - PubMed
-
- Agmon, N., W. Rettig, and C. Groth. 2002. Electronic determinants of photoacidity in cyanonaphthols. J. Am. Chem. Soc. 124:1089–1096. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

