A mitochondrial mutator system in maize
- PMID: 15681663
- PMCID: PMC1065377
- DOI: 10.1104/pp.104.053611
A mitochondrial mutator system in maize
Abstract
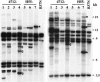
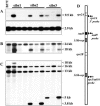
The P2 line of maize (Zea mays) is characterized by mitochondrial genome destabilization, initiated by recessive nuclear mutations. These alleles alter copy number control of mitochondrial subgenomes and disrupt normal transfer of mitochondrial genomic components to progeny, resulting in differences in mitochondrial DNA profiles among sibling plants and between parents and progeny. The mitochondrial DNA changes are often associated with variably defective phenotypes, reflecting depletion of essential mitochondrial genes. The P2 nuclear genotype can be considered a natural mutagenesis system for maize mitochondria. It dramatically accelerates mitochondrial genomic divergence by increasing low copy-number subgenomes, by rapidly amplifying aberrant recombination products, and by causing the random loss of normal components of the mitochondrial genomes.
Figures






References
-
- Bellaoui M, Martin-Canadell A, Pelletier G, Budar F (1998) Low-copy-number molecules are produced by recombination, actively maintained and can be amplified in the mitochondrial genome of Brassicaceae: relationship to reversion of the male sterile phenotype in some cybrids. Mol Gen Genet 257: 177–185 - PubMed
-
- Brown WL, Duvick DN (1958) An extreme nuclear-cytoplasmic interaction. Maize Genet Coop News Lett 32: 120–121
-
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

