Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization
- PMID: 15684028
- PMCID: PMC2171726
- DOI: 10.1083/jcb.200409091
Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization
Abstract
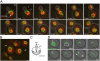
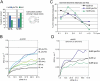
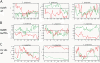
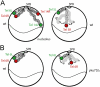
Long-range chromosome organization is known to influence nuclear function. Budding yeast centromeres cluster near the spindle pole body, whereas telomeres are grouped in five to eight perinuclear foci. Using live microscopy, we examine the relative positions of right and left telomeres of several yeast chromosomes. Integrated lac and tet operator arrays are visualized by their respective repressor fused to CFP and YFP in interphase yeast cells. The two ends of chromosomes 3 and 6 interact significantly but transiently, forming whole chromosome loops. For chromosomes 5 and 14, end-to-end interaction is less frequent, yet telomeres are closer to each other than to the centromere, suggesting that yeast chromosomes fold in a Rabl-like conformation. Disruption of telomere anchoring by deletions of YKU70 or SIR4 significantly compromises contact between two linked telomeres. These mutations do not, however, eliminate coordinated movement of telomere (Tel) 6R and Tel6L, which we propose stems from the territorial organization of yeast chromosomes.
Figures




References
-
- Belmont, A.S. 2001. Visualizing chromosome dynamics with GFP. Trends Cell Biol. 11:250–257. - PubMed
-
- Berg, H. 1993. Random Walks in Biology. Princeton University Press, Princeton, NJ. 164 pp.
-
- Bradnam, K.R., C. Seoighe, P.M. Sharp, and K.H. Wolfe. 1999. G+C content variation along and among Saccharomyces cerevisiae chromosomes. Mol. Biol. Evol. 16:666–675. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

