Mechanism for nucleoside analog-mediated abrogation of HIV-1 replication: balance between RNase H activity and nucleotide excision
- PMID: 15684061
- PMCID: PMC548555
- DOI: 10.1073/pnas.0409823102
Mechanism for nucleoside analog-mediated abrogation of HIV-1 replication: balance between RNase H activity and nucleotide excision
Abstract
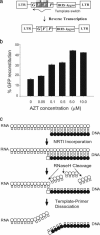
Understanding the mechanisms of HIV-1 drug resistance is critical for developing more effective antiretroviral agents and therapies. Based on our previously described dynamic copy-choice mechanism for retroviral recombination and our observations that nucleoside reverse transcriptase inhibitors (NRTIs) increase the frequency of reverse transcriptase template switching, we propose that an equilibrium exists between (i) NRTI incorporation, NRTI excision, and resumption of DNA synthesis and (ii) degradation of the RNA template by RNase H activity, leading to dissociation of the template-primer and abrogation of HIV-1 replication. As predicted by this model, mutations in the RNase H domain that reduced the rate of RNA degradation conferred high-level resistance to 3'-azido-3'-deoxythymidine and 2,3-didehydro-2,3-dideoxythymidine by as much as 180- and 10-fold, respectively, by increasing the time available for excision of incorporated NRTIs from terminated primers. These results provide insights into the mechanism by which NRTIs inhibit HIV-1 replication and imply that mutations in RNase H could significantly contribute to drug resistance either alone or in combination with NRTI-resistance mutations in reverse transcriptase.
Figures
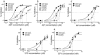


References
-
- Shafer, R. W., Winters, M. A., Palmer, S. & Merigan, T. C. (1998) Ann. Intern. Med. 128, 906-911. - PubMed
-
- Goldschmidt, V. & Marquet, R. (2004) Intern. J. Biochem. Cell Biol. 36, 1687-1705. - PubMed
-
- Arion, D., Kaushik, N., McCormick, S., Borkow, G. & Parniak, M. A. (1998) Biochemistry 37, 15908-15917. - PubMed
-
- Meyer, P. R., Matsuura, S. E., Mian, A. M., So, A. G. & Scott, W. A. (1999) Mol. Cell 4, 35-43. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

