CD8+ immunodominance among Epstein-Barr virus lytic cycle antigens directly reflects the efficiency of antigen presentation in lytically infected cells
- PMID: 15684323
- PMCID: PMC2213038
- DOI: 10.1084/jem.20041542
CD8+ immunodominance among Epstein-Barr virus lytic cycle antigens directly reflects the efficiency of antigen presentation in lytically infected cells
Abstract
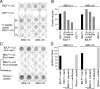
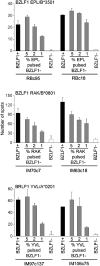
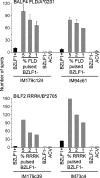
Antigen immunodominance is an unexplained feature of CD8+ T cell responses to herpesviruses, which are agents whose lytic replication involves the sequential expression of immediate early (IE), early (E), and late (L) proteins. Here, we analyze the primary CD8 response to Epstein-Barr virus (EBV) infection for reactivity to 2 IE proteins, 11 representative E proteins, and 10 representative L proteins, across a range of HLA backgrounds. Responses were consistently skewed toward epitopes in IE and a subset of E proteins, with only occasional responses to novel epitopes in L proteins. CD8+ T cell clones to representative IE, E, and L epitopes were assayed against EBV-transformed lymphoblastoid cell lines (LCLs) containing lytically infected cells. This showed direct recognition of lytically infected cells by all three sets of effectors but at markedly different levels, in the order IE > E >> L, indicating that the efficiency of epitope presentation falls dramatically with progress of the lytic cycle. Thus, EBV lytic cycle antigens display a hierarchy of immunodominance that directly reflects the efficiency of their presentation in lytically infected cells; the CD8+ T cell response thereby focuses on targets whose recognition leads to maximal biologic effect.
Figures





References
-
- Doherty, P.C., and J.P. Christensen. 2000. Accessing complexity: the dynamics of virus-specific T cell responses. Annu. Rev. Immunol. 18:561–592. - PubMed
-
- McMichael, A.J., and R.E. Phillips. 1997. Escape of human immunodeficiency virus from immune control. Annu. Rev. Immunol. 15:271–296. - PubMed
-
- Klenerman, P., V. Cerundolo, and P.R. Dunbar. 2002. Tracking T cells with tetramers: new tales from new tools. Nat. Rev. Immunol. 2:263–272. - PubMed
-
- Yewdell, J.W., and J.R. Bennink. 1999. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 17:51–58. - PubMed
-
- Cresswell, P., N. Bangia, T. Dick, and G. Diedrich. 1999. The nature of the MHC class I peptide loading complex. Immunol. Rev. 172:21–28. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

