Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function
- PMID: 15684379
- PMCID: PMC548001
- DOI: 10.1128/MCB.25.4.1258-1271.2005
Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function
Abstract
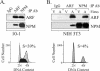
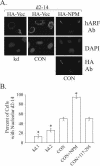
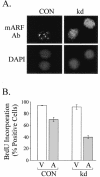
The ARF tumor suppressor is a nucleolar protein that activates p53-dependent checkpoints by binding Mdm2, a p53 antagonist. Despite persuasive evidence that ARF can bind and inactivate Mdm2 in the nucleoplasm, the prevailing view is that ARF exerts its growth-inhibitory activities from within the nucleolus. We suggest ARF primarily functions outside the nucleolus and provide evidence that it is sequestered and held inactive in that compartment by a nucleolar phosphoprotein, nucleophosmin (NPM). Most cellular ARF is bound to NPM regardless of whether cells are proliferating or growth arrested, indicating that ARF-NPM association does not correlate with growth suppression. Notably, ARF binds NPM through the same domains that mediate nucleolar localization and Mdm2 binding, suggesting that NPM could control ARF localization and compete with Mdm2 for ARF association. Indeed, NPM knockdown markedly enhanced ARF-Mdm2 association and diminished ARF nucleolar localization. Those events correlated with greater ARF-mediated growth suppression and p53 activation. Conversely, NPM overexpression antagonized ARF function while increasing its nucleolar localization. These data suggest that NPM inhibits ARF's p53-dependent activity by targeting it to nucleoli and impairing ARF-Mdm2 association.
Figures





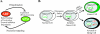
References
-
- Bertwistle, D., F. Zindy, C. J. Sherr, and M. F. Roussel. 2004. Monoclonal antibodies to the mouse p19ARF tumor suppressor protein. Hybridoma Hybridomics 23:293-300. - PubMed
-
- Biggiogera, M., K. Burki, S. H. Kaufmann, J. H. Shaper, N. Gas, F. Amalric, and S. Fakan. 1990. Nucleolar distribution of proteins B23 and nucleolin in mouse preimplantation embryos as visualized by immunoelectron microscopy. Development 110:1263-1270. - PubMed
-
- Borer, R. A., C. F. Lehner, H. M. Eppenberger, and E. A. Nigg. 1989. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 56:379-390. - PubMed
-
- Carnero, A., J. D. Hudson, C. M. Price, and D. H. Beach. 2000. p16(INK4A) and p19ARF act in overlapping pathways in cellular immortalization. Nat. Cell Biol. 2:148-155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
