Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators
- PMID: 15684387
- PMCID: PMC548005
- DOI: 10.1128/MCB.25.4.1354-1366.2005
Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators
Abstract
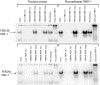
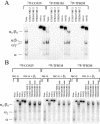
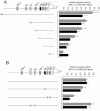
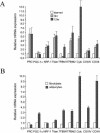
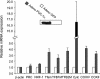
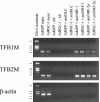
In vertebrates, mitochondrial DNA (mtDNA) transcription is initiated bidirectionally from closely spaced promoters, HSP and LSP, within the D-loop regulatory region. Early studies demonstrated that mtDNA transcription requires mitochondrial RNA polymerase and Tfam, a DNA binding stimulatory factor that is required for mtDNA maintenance. Recently, mitochondrial transcription specificity factors (TFB1M and TFB2M), which markedly enhance mtDNA transcription in the presence of Tfam and mitochondrial RNA polymerase, have been identified in mammalian cells. Here, we establish that the expression of human TFB1M and TFB2M promoters is governed by nuclear respiratory factors (NRF-1 and NRF-2), key transcription factors implicated in mitochondrial biogenesis. In addition, we show that NRF recognition sites within both TFB promoters are required for maximal trans activation by the PGC-1 family coactivators, PGC-1alpha and PRC. The physiological induction of these coactivators has been associated with the integration of NRFs and other transcription factors in a program of mitochondrial biogenesis. Finally, we demonstrate that the TFB genes are up-regulated along with Tfam and either PGC-1alpha or PRC in cellular systems where mitochondrial biogenesis is induced. Moreover, ectopic expression of PGC-1alpha is sufficient to induce the coordinate expression of all three nucleus-encoded mitochondrial transcription factors along with nuclear and mitochondrial respiratory subunits. These results support the conclusion that the coordinate regulation of nucleus-encoded mitochondrial transcription factors by NRFs and PGC-1 family coactivators is essential to the control of mitochondrial biogenesis.
Figures
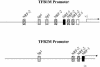




References
-
- Bogenhagen, D. F. 1996. Interaction of mtTFB and mtRNA polymerase at core promoters for transcription of Xenopus laevis mtDNA. J. Biol. Chem. 271:12036-12041. - PubMed
-
- Choi, Y. S., H. K. Lee, and Y. K. Pak. 2002. Characterization of the 5′-flanking region of the rat gene for mitochondrial transcription factor A (Tfam). Biochim. Biophys. Acta Gene Struct. Expr. 1574:200-204. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
