Reconstitution of O-specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis
- PMID: 15687196
- PMCID: PMC545620
- DOI: 10.1128/JB.187.4.1324-1333.2005
Reconstitution of O-specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis
Abstract
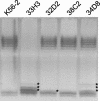
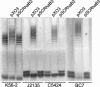
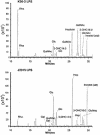
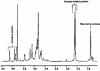
Burkholderia cenocepacia is an opportunistic bacterium that infects patients with cystic fibrosis. B. cenocepacia strains J2315, K56-2, C5424, and BC7 belong to the ET12 epidemic clone, which is transmissible among patients. We have previously shown that transposon mutants with insertions within the O antigen cluster of strain K56-2 are attenuated for survival in a rat model of lung infection. From the genomic DNA sequence of the O antigen-deficient strain J2315, we have identified an O antigen lipopolysaccharide (LPS) biosynthesis gene cluster that has an IS402 interrupting a predicted glycosyltransferase gene. A comparison with the other clonal isolates revealed that only strain K56-2, which produced O antigen and displayed serum resistance, lacked the insertion element inserted within the putative glycosyltransferase gene. We cloned the uninterrupted gene and additional flanking sequences from K56-2 and conjugated this plasmid into strains J2315, C5424, and BC7. All the exconjugants recovered the ability to form LPS O antigen. We also determined that the structure of the strain K56-2 O antigen repeat, which was absent from the LPS of strain J2315, consisted of a trisaccharide unit made of rhamnose and two N-acetylgalactosamine residues. The complexity of the gene organization of the K56-2 O antigen cluster was also investigated by reverse transcription-PCR, revealing several transcriptional units, one of which also contains genes involved in lipid A-core oligosaccharide biosynthesis.
Figures




References
-
- Aaron, S. D., W. Ferris, D. A. Henry, D. P. Speert, and N. E. Macdonald. 2000. Multiple combination bactericidal antibiotic testing for patients with cystic fibrosis infected with Burkholderia cepacia. Am. J. Respir. Crit. Care Med. 161:1206-1212. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Baird, R. M., H. Brown, A. W. Smith, and M. L. Watson. 1999. Burkholderia cepacia is resistant to the antimicrobial activity of airway epithelial cells. Immunopharmacology 44:267-272. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

