Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum
- PMID: 15687204
- PMCID: PMC545617
- DOI: 10.1128/JB.187.4.1392-1404.2005
Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum
Abstract
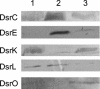
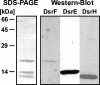
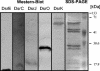
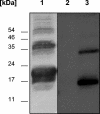
Seven new genes designated dsrLJOPNSR were identified immediately downstream of dsrABEFHCMK, completing the dsr gene cluster of the phototrophic sulfur bacterium Allochromatium vinosum D (DSM 180(T)). Interposon mutagenesis proved an essential role of the encoded proteins for the oxidation of intracellular sulfur, an obligate intermediate during the oxidation of sulfide and thiosulfate. While dsrR and dsrS encode cytoplasmic proteins of unknown function, the other genes encode a predicted NADPH:acceptor oxidoreductase (DsrL), a triheme c-type cytochrome (DsrJ), a periplasmic iron-sulfur protein (DsrO), and an integral membrane protein (DsrP). DsrN resembles cobyrinic acid a,c-diamide synthases and is probably involved in the biosynthesis of siro(heme)amide, the prosthetic group of the dsrAB-encoded sulfite reductase. The presence of most predicted Dsr proteins in A. vinosum was verified by Western blot analysis. With the exception of the constitutively present DsrC, the formation of Dsr gene products was greatly enhanced by sulfide. DsrEFH were purified from the soluble fraction and constitute a soluble alpha(2)beta(2)gamma(2)-structured 75-kDa holoprotein. DsrKJO were purified from membranes pointing at the presence of a transmembrane electron-transporting complex consisting of DsrKMJOP. In accordance with the suggestion that related complexes from dissimilatory sulfate reducers transfer electrons to sulfite reductase, the A. vinosum Dsr complex is copurified with sulfite reductase, DsrEFH, and DsrC. We therefore now have an ideal and unique possibility to study the interaction of sulfite reductase with other proteins and to clarify the long-standing problem of electron transport from and to sulfite reductase, not only in phototrophic bacteria but also in sulfate-reducing prokaryotes.
Figures


References
-
- Arslan, E., H. Schulz, R. Zufferey, P. Kunzler, and L. Thöny-Meyer. 1998. Overproduction of Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochem. Biophys. Res. Commun. 251:744-747. - PubMed
-
- Ausubel, F. A., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1997. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
-
- Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors. Mol. Microbiol. 22:393-404. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

