Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells
- PMID: 15687256
- PMCID: PMC546510
- DOI: 10.1101/gad.1254305
Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells
Erratum in
- Genes Dev. 2008 Jun 15;22(12):1717. Sayegh, Camil [corrected to Sayegh, Camil E]
Abstract
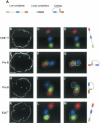
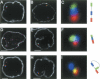
The immunoglobulin heavy-chain (IgH) locus undergoes large-scale contraction in B cells poised to undergo IgH V(D)J recombination. We considered the possibility that looping of distinct IgH V regions plays a role in promoting long-range interactions. Here, we simultaneously visualize three subregions of the IgH locus, using three-dimensional fluorescence in situ hybridization. Looping within the IgH locus was observed in both B- and T-lineage cells. However, monoallelic looping of IgH V regions into close proximity of the IgH DJ cluster was detected in developing B cells with significantly higher frequency when compared with hematopoietic progenitor or CD8+ T-lineage cells. Looping of a subset of IgH V regions, albeit at lower frequency, was also observed in RAG-deficient pro-B cells. Based on these observations, we propose that Ig loci are repositioned by a looping mechanism prior to IgH V(D)J rearrangement to facilitate the joining of Ig variable, diversity, and joining segments.
Figures
References
-
- Baxter J., Merkenschlager, M., and Fisher, A.G. 2002. Nuclear organisation and gene expression. Curr. Opin. Cell Biol. 14: 372-376. - PubMed
-
- Bergman Y. and Cedar, H. 2004. A stepwise epigenetic process controls immunoglobulin allelic exclusion. Nat. Rev. Immunol. 4: 753-761. - PubMed
-
- Carter D., Chakalova, L., Osborne, C.S., Dai, Y.F., and Fraser, P. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32: 623-626. - PubMed
-
- Chevillard C., Ozaki, J., Herring, C.D., and Riblet, R. 2002. A three-megabase yeast artificial chromosome contig spanning the C57BL mouse Igh locus. J. Immunol. 168: 5659-5666. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
