Evolution of recombination due to random drift
- PMID: 15687279
- PMCID: PMC1449609
- DOI: 10.1534/genetics.104.032821
Evolution of recombination due to random drift
Abstract
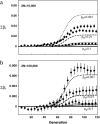
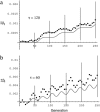
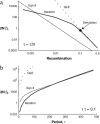
In finite populations subject to selection, genetic drift generates negative linkage disequilibrium, on average, even if selection acts independently (i.e., multiplicatively) upon all loci. Negative disequilibrium reduces the variance in fitness and hence, by Fisher's (1930) fundamental theorem, slows the rate of increase in mean fitness. Modifiers that increase recombination eliminate the negative disequilibria that impede selection and consequently increase in frequency by "hitchhiking." Thus, stochastic fluctuations in linkage disequilibrium in finite populations favor the evolution of increased rates of recombination, even in the absence of epistatic interactions among loci and even when disequilibrium is initially absent. The method developed within this article allows us to quantify the strength of selection acting on a modifier allele that increases recombination in a finite population. The analysis indicates that stochastically generated linkage disequilibria do select for increased recombination, a result that is confirmed by Monte Carlo simulations. Selection for a modifier that increases recombination is highest when linkage among loci is tight, when beneficial alleles rise from low to high frequency, and when the population size is small.
Figures
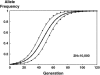
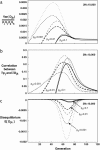
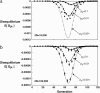

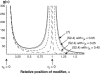

References
-
- Barton, N. H., 1995. a A general model for the evolution of recombination. Genet. Res. 65: 123–144. - PubMed
-
- Barton, N. H., 1998. The effect of hitch-hiking on neutral genealogies. Genet. Res. 72: 123–133.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

