Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain
- PMID: 15689398
- PMCID: PMC549489
- DOI: 10.1073/pnas.0409853102
Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain
Abstract
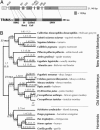
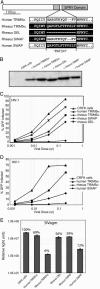
Primate genomes encode a variety of innate immune strategies to defend themselves against retroviruses. One of these, TRIM5alpha, can restrict diverse retroviruses in a species-specific manner. Thus, whereas rhesus TRIM5alpha can strongly restrict HIV-1, human TRIM5alpha only has weak HIV-1 restriction. The biology of TRIM5alpha restriction suggests that it is locked in an antagonistic conflict with the proteins encoding the viral capsid. Such antagonistic interactions frequently result in rapid amino acid replacements at the protein-protein interface, as each genetic entity vies for evolutionary dominance. By analyzing its evolutionary history, we find strong evidence for ancient positive selection in the primate TRIM5alpha gene. This selection is strikingly variable with some of the strongest selection occurring in the human lineage. This history suggests that TRIM5alpha evolution has been driven by antagonistic interactions with a wide variety of viruses and endogenous retroviruses that predate the origin of primate lentiviruses. A 13-aa "patch" in the SPRY protein domain bears a dense concentration of positively selected residues, potentially implicating it as an antiviral interface. By using functional studies of chimeric TRIM5alpha genes, we show that this patch is generally essential for retroviral restriction and is responsible for most of the species-specific antiretroviral restriction activity. Our study highlights the power of evolutionary analyses, in which positive selection identifies not only the age of genetic conflict but also the interaction interface where this conflict plays out.
Figures

Comment in
-
The power of phylogenetic comparison in revealing protein function.Proc Natl Acad Sci U S A. 2005 Mar 1;102(9):3179-80. doi: 10.1073/pnas.0500371102. Epub 2005 Feb 22. Proc Natl Acad Sci U S A. 2005. PMID: 15728394 Free PMC article. No abstract available.
References
-
- Sheehy, A. M., Gaddis, N. C., Choi, J. D. & Malim, M. H. (2002) Nature 418, 646–650. - PubMed
-
- Stremlau, M., Owens, C. M., Perron, M. J., Kiessling, M., Autissier, P. & Sodroski, J. (2004) Nature 427, 848–853. - PubMed
-
- Xu, L., Yang, L., Moitra, P. K., Hashimoto, K., Rallabhandi, P., Kaul, S., Meroni, G., Jensen, J. P., Weissman, A. M. & D'Arpa, P. (2003) Exp. Cell Res. 288, 84–93. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

