HIV-1 clade promoters strongly influence spatial and temporal dynamics of viral replication in vivo
- PMID: 15690084
- PMCID: PMC546425
- DOI: 10.1172/JCI22873
HIV-1 clade promoters strongly influence spatial and temporal dynamics of viral replication in vivo
Abstract
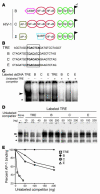
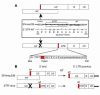
Although the primary determinant of cell tropism is the interaction of viral envelope or capsid proteins with cellular receptors, other viral elements can strongly modulate viral replication. While the HIV-1 promoter is polymorphic for a variety of transcription factor binding sites, the impact of these polymorphisms on viral replication in vivo is not known. To address this issue, we engineered isogenic SIVmac239 chimeras harboring the core promoter/enhancer from HIV-1 clades B, C, and E. Here it is shown that the clade C and E core promoters/enhancers bear a noncanonical activator protein-1 (AP-1) binding site, absent from the corresponding clade B region. Relative ex vivo replication of chimeras was strongly dependent on the tissue culture system used. Notably, in thymic histocultures, replication of the clade C chimera was favored by IL-7 enrichment, which suggests that the clade C polymorphism in the AP-1 and NF-kappaB binding sites is involved. Simultaneous infection of rhesus macaques with the 3 chimeras revealed a strong predominance of the clade C chimera during primary infection. Thereafter, the B chimera dominated in all tissues. These data show that the clade C promoter is particularly adapted to sustain viral replication in primary viremia and that clade-specific promoter polymorphisms constitute a major determinant for viral replication.
Figures
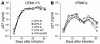


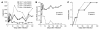

References
-
- Collman RG, Perno CF, Crowe SM, Stevenson M, Montaner LJ. HIV and cells of macrophage/dendritic lineage and other non-T cell reservoirs: new answers yield new questions. J. Leukoc. Biol. 2003;74:631–634. - PubMed
-
- Pierson TC, Doms RW. HIV-1 entry and its inhibition. Curr. Top. Microbiol. Immunol. 2003;281:1–27. - PubMed
-
- Roof P, et al. Differential regulation of HIV-1 clade-specific B, C, and E long terminal repeats by NF-kappaB and the Tat transactivator. Virology. 2002;296:77–83. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

