Molecular modeling and chemical modification for finding peptide inhibitor against severe acute respiratory syndrome coronavirus main proteinase
- PMID: 15691506
- PMCID: PMC7094278
- DOI: 10.1016/j.ab.2004.10.003
Molecular modeling and chemical modification for finding peptide inhibitor against severe acute respiratory syndrome coronavirus main proteinase
Abstract
Severe acute respiratory syndrome (SARS) is a respiratory disease caused by a newly found virus, called SARS coronavirus. In this study, the cleavage mechanism of the SARS coronavirus main proteinase (Mpro or 3CLpro) on the octapeptide NH2-AVLQ downward arrowSGFR-COOH was investigated using molecular mechanics and quantum mechanics simulations based on the experimental structure of the proteinase. It has been observed that the catalytic dyad (His-41/Cys-145) site between domains I and II attracts the pi electron density from the peptide bond Gln-Ser, increasing the positive charge on C(CO) of Gln and the negative charge on N(NH) of Ser, so as to weaken the Gln-Ser peptide bond. The catalytic functional group is the imidazole group of His-41 and the S in Cys-145. Ndelta1 on the imidazole ring plays the acid-base catalytic role. Based on the "distorted key theory" [K.C. Chou, Anal. Biochem. 233 (1996) 1-14], the possibility to convert the octapeptide to a competent inhibitor has been studied. It has been found that the chemical bond between Gln and Ser will become much stronger and no longer cleavable by the SARS enzyme after either changing the carbonyl group CO of Gln to CH2 or CF2 or changing the NH of Ser to CH2 or CF2. The octapeptide thus modified might become an effective inhibitor or a potential drug candidate against SARS.
Figures
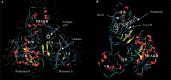
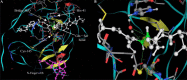
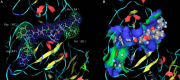
Similar articles
-
Polyprotein cleavage mechanism of SARS CoV Mpro and chemical modification of the octapeptide.Peptides. 2004 Nov;25(11):1857-64. doi: 10.1016/j.peptides.2004.06.018. Peptides. 2004. PMID: 15501516 Free PMC article.
-
Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs.Science. 2003 Jun 13;300(5626):1763-7. doi: 10.1126/science.1085658. Epub 2003 May 13. Science. 2003. PMID: 12746549
-
Application of bioinformatics in search for cleavable peptides of SARS-CoV M(pro) and chemical modification of octapeptides.Med Chem. 2005 May;1(3):209-13. doi: 10.2174/1573406053765468. Med Chem. 2005. PMID: 16787316
-
Drug targets in severe acute respiratory syndrome (SARS) virus and other coronavirus infections.Infect Disord Drug Targets. 2009 Apr;9(2):223-45. doi: 10.2174/187152609787847659. Infect Disord Drug Targets. 2009. PMID: 19275708 Review.
-
Recent developments in the virology and antiviral research of severe acute respiratory syndrome coronavirus.Infect Disord Drug Targets. 2007 Mar;7(1):29-41. doi: 10.2174/187152607780090739. Infect Disord Drug Targets. 2007. PMID: 17346209 Review.
Cited by
-
Computational studies of the binding modes of A 2A adenosine receptor antagonists.Amino Acids. 2008 Aug;35(2):389-96. doi: 10.1007/s00726-007-0604-2. Epub 2007 Nov 5. Amino Acids. 2008. PMID: 17978889 Free PMC article.
-
iEzy-drug: a web server for identifying the interaction between enzymes and drugs in cellular networking.Biomed Res Int. 2013;2013:701317. doi: 10.1155/2013/701317. Epub 2013 Nov 26. Biomed Res Int. 2013. PMID: 24371828 Free PMC article.
-
Peptide-Based Inhibitors for SARS-CoV-2 and SARS-CoV.Adv Ther (Weinh). 2021 Oct;4(10):2100104. doi: 10.1002/adtp.202100104. Epub 2021 Aug 6. Adv Ther (Weinh). 2021. PMID: 34514085 Free PMC article. Review.
-
Unraveling the origin of interactions of hydroxychloroquine with the receptor-binding domain of SARS-CoV-2 in aqueous medium.Chem Phys Lett. 2021 Feb;764:138280. doi: 10.1016/j.cplett.2020.138280. Epub 2020 Dec 19. Chem Phys Lett. 2021. PMID: 33362291 Free PMC article.
-
A classification of countries and regions by degree of the spread of coronavirus based on statistical criteria.Expert Syst Appl. 2021 Jun 15;172:114654. doi: 10.1016/j.eswa.2021.114654. Epub 2021 Feb 1. Expert Syst Appl. 2021. PMID: 33551577 Free PMC article.
References
-
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
-
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
-
- Shortridge K.F. Severe acute respiratory syndrome and influenza: virus incursions from southern China. Am. J. Respir. Crit. Care Med. 2003;168:1416–1420. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

