Structure of sediment-associated microbial communities along a heavy-metal contamination gradient in the marine environment
- PMID: 15691917
- PMCID: PMC546797
- DOI: 10.1128/AEM.71.2.679-690.2005
Structure of sediment-associated microbial communities along a heavy-metal contamination gradient in the marine environment
Abstract
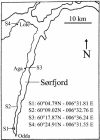
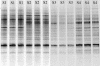
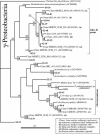
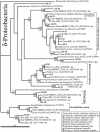
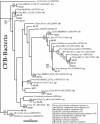
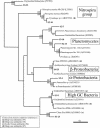
Microbial community composition and structure were characterized in marine sediments contaminated for >80 years with cadmium, copper, lead, and zinc. Four sampling sites that encompass a wide range of sediment metal loads were compared in a Norwegian fjord (Sorfjord). HCl-extractable metals and organic matter constantly decreased from the most contaminated site (S1) to the control site (S4). All sampling sites presented low polychlorinated biphenyl (PCB) concentrations (Sigma(7)PCB < 7.0 ng g [dry weight](-1)). The biomass ranged from 4.3 x 10(8) to 13.4 x 10(8) cells g (dry weight) of sediments(-1) and was not correlated to metal levels. Denaturing gradient gel electrophoresis indicated that diversity was not affected by the contamination. The majority of the partial 16S rRNA sequences obtained were classified in the gamma- and delta-Proteobacteria and in the Cytophaga-Flexibacter-Bacteroides (CFB) bacteria. Some sequences were closely related to other sequences from polluted marine sediments. The abundances of seven phylogenetic groups were determined by using fluorescent in situ hybridization (FISH). FISH was impaired in S1 by high levels of autofluorescing particles. For S2 to S4, the results indicated that the HCl-extractable Cu, Pb, and Zn were negatively correlated with the abundance of gamma-Proteobacteria and CFB bacteria. delta-Proteobacteria were not correlated with HCl-extractable metals. Bacteria of the Desulfosarcina-Desulfococcus group were detected in every site and represented 6 to 14% of the DAPI (4',6'-diamidino-2-phenylindole) counts. Although factors other than metals may explain the distribution observed, the information presented here may be useful in predicting long-term effects of heavy-metal contamination in the marine environment.
Figures

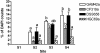
References
-
- Austin, B. 1989. Methods in aquatic bacteriology. John Wiley & Sons, Chichester, United Kingdom.
-
- Bååth, E. 1989. Effects of heavy metals in soil on microbial processes and populations. Water Air Soil Pollut. 47:335-379.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

