Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria
- PMID: 15691937
- PMCID: PMC546801
- DOI: 10.1128/AEM.71.2.826-834.2005
Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria
Abstract
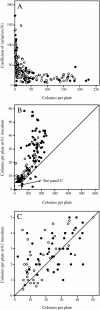
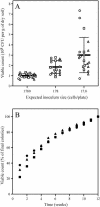
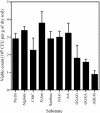
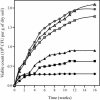
Soils are inhabited by many bacteria from phylogenetic groups that are poorly studied because representatives are rarely isolated in cultivation studies. Part of the reason for the failure to cultivate these bacteria is the low frequency with which bacterial cells in soil form visible colonies when inoculated onto standard microbiological media, resulting in low viable counts. We investigated the effects of three factors on viable counts, assessed as numbers of CFU on solid media, and on the phylogenetic groups to which the isolated colony-forming bacteria belong. These factors were inoculum size, growth medium, and incubation time. Decreasing the inoculum size resulted in significant increases in the viable count but did not appear to affect colony formation by members of rarely isolated groups. Some media that are traditionally used for soil microbiological studies returned low viable counts and did not result in the isolation of members of rarely isolated groups. Newly developed media, in contrast, resulted in high viable counts and in the isolation of many members of rarely isolated groups, regardless of the inoculum size. Increased incubation times of up to 3 months allowed the development of visible colonies of members of rarely isolated groups in conjunction with the use of appropriate media. Once isolated, pure cultures of members of rarely isolated groups took longer to form visible colonies than did members of commonly isolated groups. Using these new media and extended incubation times, we were able to isolate many members of the phyla Acidobacteria (subdivisions 1, 2, 3, and 4), Gemmatimonadetes, Chloroflexi, and Planctomycetes (including representatives of the previously uncultured WPS-1 lineage) as well as members of the subclasses Rubrobacteridae and Acidimicrobidae of the phylum Actinobacteria.
Figures
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Balestra, G. M., and I. J. Misaghi. 1997. Increasing the efficiency of the plate count method for estimating bacterial diversity. J. Microbiol. Methods 30:111-117.
-
- Buckley, D. H., and T. M. Schmidt. 2002. Exploring the biodiversity of soil—a microbial rain forest, p. 183-208. In J. T. Staley and A.-L. Reysenbach (ed.), Biodiversity of microbial life: foundation of the Earth's biosphere. Wiley-Liss, Inc., New York, N.Y.
-
- Casida, L. E., Jr. 1968. Methods for the isolation and estimation of activity of soil bacteria, p. 97-122. In T. R. G. Gray and D. Parkinson (ed.), The ecology of soil bacteria. Liverpool University Press, Liverpool, United Kingdom.
-
- Conn, H. J. 1918. The microscopic study of bacteria and fungi in soil. N. Y. Agric. Exp. Stn. Tech. Bull. 64:3-20.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

