Detection and discrimination of Cryptosporidium parvum and C. hominis in water samples by immunomagnetic separation-PCR
- PMID: 15691946
- PMCID: PMC546695
- DOI: 10.1128/AEM.71.2.898-903.2005
Detection and discrimination of Cryptosporidium parvum and C. hominis in water samples by immunomagnetic separation-PCR
Abstract
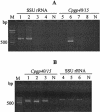
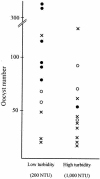
Cryptosporidium parvum and C. hominis have been the cause of large and serious outbreaks of waterborne cryptosporidiosis. A specific and sensitive recovery-detection method is required for control of this pathogen in drinking water. In the present study, nested PCR-restriction fragment length polymorphism (RFLP), which targets the divergent Cpgp40/15 gene, was developed. This nested PCR detected only the gene derived from C. parvum and C. hominis strains, and RFLP was able to discriminate between the PCR products from C. parvum and C. hominis. To evaluate the sensitivity of nested PCR, C. parvum oocysts inoculated in water samples of two different turbidities were recovered by immunomagnetic separation (IMS) and detected by nested PCR and fluorescent antibody assay (FA). Genetic detection by nested PCR and oocyst number confirmed by FA were compared, and the results suggested that detection by nested PCR depends on the confirmed oocyst number and that nested PCR in combination with IMS has the ability to detect a single oocyst in a water sample. We applied an agitation procedure with river water solids to which oocysts were added to evaluate the recovery and detection by the procedure in environmental samples and found some decrease in the rate of detection by IMS.
Figures
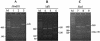
References
-
- DuPont, H. L., C. L. Chappell, C. R. Sterling, P. C. Okhuysen, J. B. Rose, and W. Jakubowski. 1995. The infectivity of Cryptosporidium parvum in healthy volunteers. N. Engl. J. Med. 332:855-859. - PubMed
-
- Ijzerman, M. M., D. R. Dahling, and G. S. Fout. 1997. A method to remove environmental inhibitors prior to the detection of waterborne enteric viruses by reverse transcription-polymerase chain reaction. J. Virol. Methods 63:145-153. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

