Phylogenetic characterization of 16S rRNA gene clones from deep-groundwater microorganisms that pass through 0.2-micrometer-pore-size filters
- PMID: 15691970
- PMCID: PMC546738
- DOI: 10.1128/AEM.71.2.1084-1088.2005
Phylogenetic characterization of 16S rRNA gene clones from deep-groundwater microorganisms that pass through 0.2-micrometer-pore-size filters
Abstract
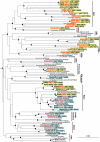
A total of 247 clones of 16S rRNA genes from microorganisms captured by 0.2- and 0.1-microm-pore-size filters from sedimentary and granite rock aquifers were amplified and yielded 37 operational taxonomic units (OTUs). Fifteen OTUs captured by 0.1-microm-pore-size filters were affiliated with the candidate divisions OD1 and OP11, representing novel lineages. On the other hand, OTUs captured by 0.2-microm-pore-size filters were largely affiliated with Betaproteobacteria.
Figures
References
-
- Åkerman, K. K., I. Kuronen, and E. O. Kajander. 1993. Scanning electron microscopy of nanobacteria—novel biofilm producing organisms in blood. Scanning 15(Suppl.):90-91.
-
- Elsaied, H. E., M. Sato, and T. Naganuma. 2001. Viable Cytophaga-like bacterium in the 0.2 μm-filtrate seawater. Syst. Appl. Microbiol. 24:0-5. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

