High-level acetaldehyde production in Lactococcus lactis by metabolic engineering
- PMID: 15691976
- PMCID: PMC546684
- DOI: 10.1128/AEM.71.2.1109-1113.2005
High-level acetaldehyde production in Lactococcus lactis by metabolic engineering
Abstract
Efficient conversion of glucose to acetaldehyde is achieved by nisin-controlled overexpression of Zymomonas mobilis pyruvate decarboxylase (pdc) and Lactococcus lactis NADH oxidase (nox) in L. lactis. In resting cells, almost 50% of the glucose consumed could be redirected towards acetaldehyde by combined overexpression of pdc and nox under anaerobic conditions.
Figures
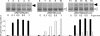
References
-
- Bongers, R. S., M. H. N. Hoefnagel, M. J. C. Starrenburg, M. A. J. Siemerink, J. G. A. Arends, J. Hugenholtz, and M. Kleerebezem. 2003. IS981-mediated adaptive evolution recovers lactate production by ldhB transcription activation in a lactate dehydrogenase-deficient strain of Lactococcus lactis. J. Bacteriol. 185:4499-4507. - PMC - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:52-58. - PubMed
-
- Bringer-Meyer, S., K. L. Schminz, and H. Sahm. 1986. Pyruvate decarboxylase from Zymomonas mobilis. Isolation and partial characterization. Arch. Microbiol. 146:105-110.
-
- Casabadan, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

