The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription
- PMID: 15692559
- PMCID: PMC554125
- DOI: 10.1038/sj.emboj.7600575
The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription
Abstract
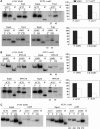
Transcription and processing of mRNA precursors are coordinated events that require numerous complex interactions to ensure that they are successfully executed. We described previously an unexpected association between a transcription factor, PC4 (or Sub1 in yeast), and an mRNA polyadenylation factor, CstF-64 (Rna15 in yeast), and provided evidence that this was important for efficient transcription elongation. Here we provide insight into the mechanism by which this occurs. We show that Sub1 and Rna15 are recruited to promoters and present along the length of several yeast genes. Allele-specific genetic interactions between SUB1 and genes encoding an RNA polymerase II (RNAP II)-specific kinase (KIN28) and phosphatase (FCP1) suggest that Sub1 influences and/or is sensitive to the phosphorylation status of elongating RNAP II. Remarkably, we find that cells lacking Sub1 display decreased accumulation of Fcp1, altered RNAP II phosphorylation and decreased crosslinking of RNAP II to transcribed genes. Our data provide evidence that Rna15 and Sub1 are present along the length of several genes and that Sub1 facilitates elongation by influencing enzymes that modify RNAP II.
Figures





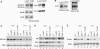
Similar articles
-
Sub1 globally regulates RNA polymerase II C-terminal domain phosphorylation.Mol Cell Biol. 2010 Nov;30(21):5180-93. doi: 10.1128/MCB.00819-10. Epub 2010 Sep 7. Mol Cell Biol. 2010. PMID: 20823273 Free PMC article.
-
Sfl1 functions via the co-repressor Ssn6-Tup1 and the cAMP-dependent protein kinase Tpk2.J Mol Biol. 2001 Jun 22;309(5):1007-15. doi: 10.1006/jmbi.2001.4742. J Mol Biol. 2001. PMID: 11399075
-
Regulation of elongating RNA polymerase II by forkhead transcription factors in yeast.Science. 2003 Apr 18;300(5618):492-5. doi: 10.1126/science.1081379. Science. 2003. PMID: 12702877
-
Sub1/PC4, a multifaceted factor: from transcription to genome stability.Curr Genet. 2017 Dec;63(6):1023-1035. doi: 10.1007/s00294-017-0715-6. Epub 2017 May 31. Curr Genet. 2017. PMID: 28567479 Review.
-
Sub1/PC4 a chromatin associated protein with multiple functions in transcription.RNA Biol. 2010 May-Jun;7(3):287-90. doi: 10.4161/rna.7.3.11491. Epub 2010 May 9. RNA Biol. 2010. PMID: 20305379 Review.
Cited by
-
Sub1 functions in osmoregulation and in transcription by both RNA polymerases II and III.Mol Cell Biol. 2009 Apr;29(8):2308-21. doi: 10.1128/MCB.01841-08. Epub 2009 Feb 9. Mol Cell Biol. 2009. PMID: 19204085 Free PMC article.
-
Transcriptional activators enhance polyadenylation of mRNA precursors.Mol Cell. 2011 Feb 18;41(4):409-18. doi: 10.1016/j.molcel.2011.01.022. Mol Cell. 2011. PMID: 21329879 Free PMC article.
-
AtCyp59 is a multidomain cyclophilin from Arabidopsis thaliana that interacts with SR proteins and the C-terminal domain of the RNA polymerase II.RNA. 2006 Apr;12(4):631-43. doi: 10.1261/rna.2226106. Epub 2006 Feb 22. RNA. 2006. PMID: 16497658 Free PMC article.
-
Enhancers regulate 3' end processing activity to control expression of alternative 3'UTR isoforms.Nat Commun. 2022 May 17;13(1):2709. doi: 10.1038/s41467-022-30525-y. Nat Commun. 2022. PMID: 35581194 Free PMC article.
-
Context-specific regulation and function of mRNA alternative polyadenylation.Nat Rev Mol Cell Biol. 2022 Dec;23(12):779-796. doi: 10.1038/s41580-022-00507-5. Epub 2022 Jul 7. Nat Rev Mol Cell Biol. 2022. PMID: 35798852 Free PMC article. Review.
References
-
- Ahn SH, Kim M, Buratowski S (2004) Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13: 67–76 - PubMed
-
- Aranda A, Proudfoot NJ (2001) Transcriptional termination factors for RNA polymerase II in yeast. Mol Cell 7: 1003–1011 - PubMed
-
- Bensaude O, Bonnet F, Casse C, Dubois MF, Nguyen VT, Palancade B (1999) Regulated phosphorylation of the RNA polymerase II C-terminal domain (CTD). Biochem Cell Biol 77: 249–255 - PubMed
-
- Bloch JC, Perrin F, Lacroute F (1978) Yeast temperature-sensitive mutants specifically impaired in processing of poly(A)-containing RNAs. Mol Gen Genet 165: 123–127 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

