Comparison of the oxidative phosphorylation (OXPHOS) nuclear genes in the genomes of Drosophila melanogaster, Drosophila pseudoobscura and Anopheles gambiae
- PMID: 15693940
- PMCID: PMC551531
- DOI: 10.1186/gb-2005-6-2-r11
Comparison of the oxidative phosphorylation (OXPHOS) nuclear genes in the genomes of Drosophila melanogaster, Drosophila pseudoobscura and Anopheles gambiae
Abstract
Background: In eukaryotic cells, oxidative phosphorylation (OXPHOS) uses the products of both nuclear and mitochondrial genes to generate cellular ATP. Interspecies comparative analysis of these genes, which appear to be under strong functional constraints, may shed light on the evolutionary mechanisms that act on a set of genes correlated by function and subcellular localization of their products.
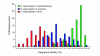
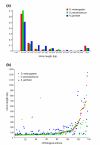
Results: We have identified and annotated the Drosophila melanogaster, D. pseudoobscura and Anopheles gambiae orthologs of 78 nuclear genes encoding mitochondrial proteins involved in oxidative phosphorylation by a comparative analysis of their genomic sequences and organization. We have also identified 47 genes in these three dipteran species each of which shares significant sequence homology with one of the above-mentioned OXPHOS orthologs, and which are likely to have originated by duplication during evolution. Gene structure and intron length are essentially conserved in the three species, although gain or loss of introns is common in A. gambiae. In most tissues of D. melanogaster and A. gambiae the expression level of the duplicate gene is much lower than that of the original gene, and in D. melanogaster at least, its expression is almost always strongly testis-biased, in contrast to the soma-biased expression of the parent gene.
Conclusions: Quickly achieving an expression pattern different from the parent genes may be required for new OXPHOS gene duplicates to be maintained in the genome. This may be a general evolutionary mechanism for originating phenotypic changes that could lead to species differentiation.
Figures

Similar articles
-
The MitoDrome database annotates and compares the OXPHOS nuclear genes of Drosophila melanogaster, Drosophila pseudoobscura and Anopheles gambiae.Mitochondrion. 2006 Oct;6(5):252-7. doi: 10.1016/j.mito.2006.07.001. Epub 2006 Jul 21. Mitochondrion. 2006. PMID: 16982216
-
Comparison of K+-channel genes within the genomes of Anopheles gambiae and Drosophila melanogaster.Genome Biol. 2003;4(9):R58. doi: 10.1186/gb-2003-4-9-r58. Epub 2003 Aug 20. Genome Biol. 2003. PMID: 12952537 Free PMC article.
-
MitoRes: a resource of nuclear-encoded mitochondrial genes and their products in Metazoa.BMC Bioinformatics. 2006 Jan 24;7:36. doi: 10.1186/1471-2105-7-36. BMC Bioinformatics. 2006. PMID: 16433928 Free PMC article.
-
Identification of the Anopheles gambiae ATP-binding cassette transporter superfamily genes.Mol Cells. 2003 Apr 30;15(2):150-8. Mol Cells. 2003. PMID: 12803476 Review.
-
Metazoan OXPHOS gene families: evolutionary forces at the level of mitochondrial and nuclear genomes.Biochim Biophys Acta. 2006 Sep-Oct;1757(9-10):1171-8. doi: 10.1016/j.bbabio.2006.04.021. Epub 2006 May 4. Biochim Biophys Acta. 2006. PMID: 16781661 Review.
Cited by
-
Mitochondrial-Nuclear Interactions Mediate Sex-Specific Transcriptional Profiles in Drosophila.Genetics. 2016 Oct;204(2):613-630. doi: 10.1534/genetics.116.192328. Epub 2016 Aug 24. Genetics. 2016. PMID: 27558138 Free PMC article.
-
Genome-Wide Analysis Suggests the Relaxed Purifying Selection Affect the Evolution of WOX Genes in Pyrus bretschneideri, Prunus persica, Prunus mume, and Fragaria vesca.Front Genet. 2017 Jun 15;8:78. doi: 10.3389/fgene.2017.00078. eCollection 2017. Front Genet. 2017. PMID: 28663757 Free PMC article.
-
High-resolution transcriptional profiling of Anopheles gambiae spermatogenesis reveals mechanisms of sex chromosome regulation.Sci Rep. 2019 Oct 16;9(1):14841. doi: 10.1038/s41598-019-51181-1. Sci Rep. 2019. PMID: 31619757 Free PMC article.
-
Drcd-1 related: a positively selected spermatogenesis retrogene in Drosophila.Genetica. 2010 Oct;138(9-10):925-37. doi: 10.1007/s10709-010-9474-8. Epub 2010 Aug 8. Genetica. 2010. PMID: 20694743 Free PMC article.
-
Rapid evolution of mitochondrion-related genes in haplodiploid arthropods.BMC Biol. 2024 Oct 10;22(1):229. doi: 10.1186/s12915-024-02027-4. BMC Biol. 2024. PMID: 39390511 Free PMC article.
References
-
- Bergman CM, Pfeiffer BD, Rincon-Limas DE, Hoskins RA, Gnirke A, Mungall CJ, Wang AM, Kronmiller B, Pacleb J, Park S, et al. Assessing the impact of comparative genomic sequence data on the functional annotation of the Drosophila genome. Genome Biol. 2002;3:research0086.1–0086.20. doi: 10.1186/gb-2002-3-12-research0086. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

