Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation
- PMID: 15695584
- PMCID: PMC548991
- DOI: 10.1073/pnas.0409474102
Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation
Abstract
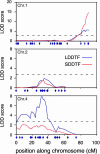
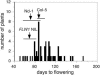
Much of the flowering time variation in wild strains of Arabidopsis thaliana is due to allelic variation at two epistatically acting loci, FRIGIDA (FRI) and FLOWERING LOCUS C (FLC). FLC encodes a MADS (MCM1/AGAMOUS/DEFICIENS/SRF1) domain transcription factor that directly represses a series of flowering-promoting genes. FRI and FLC, however, do not explain all of the observed variation, especially when plants are grown in short days. To identify loci that act in addition to FRI and FLC in controlling flowering of natural accessions, we have analyzed a recombinant inbred line population derived from crosses of accession Niederzenz (Nd) to Columbia, both of which contain natural FRI lesions. Quantitative trait locus mapping and genomic DNA analysis by microarray hybridization were used to identify candidate genes affecting variation in flowering behavior. In both long and short days, the quantitative trait locus of largest effect, termed FLOWERING 1 (FLW1), was found to be associated with a Nd-specific deletion of FLOWERING LOCUS M (FLM), which encodes a floral repressor closely related to FLC. Analysis of near isogenic lines and quantitative transgenic complementation experiments confirmed that the FLM deletion is, in large part, responsible for the early flowering of the Nd accession.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

